Xiaoyan Li
RPC-Bench: A Fine-grained Benchmark for Research Paper Comprehension
Jan 14, 2026Abstract:Understanding research papers remains challenging for foundation models due to specialized scientific discourse and complex figures and tables, yet existing benchmarks offer limited fine-grained evaluation at scale. To address this gap, we introduce RPC-Bench, a large-scale question-answering benchmark built from review-rebuttal exchanges of high-quality computer science papers, containing 15K human-verified QA pairs. We design a fine-grained taxonomy aligned with the scientific research flow to assess models' ability to understand and answer why, what, and how questions in scholarly contexts. We also define an elaborate LLM-human interaction annotation framework to support large-scale labeling and quality control. Following the LLM-as-a-Judge paradigm, we develop a scalable framework that evaluates models on correctness-completeness and conciseness, with high agreement to human judgment. Experiments reveal that even the strongest models (GPT-5) achieve only 68.2% correctness-completeness, dropping to 37.46% after conciseness adjustment, highlighting substantial gaps in precise academic paper understanding. Our code and data are available at https://rpc-bench.github.io/.
Beyond 1D: Vision Transformers and Multichannel Signal Images for PPG-to-ECG Reconstruction
May 27, 2025Abstract:Reconstructing ECG from PPG is a promising yet challenging task. While recent advancements in generative models have significantly improved ECG reconstruction, accurately capturing fine-grained waveform features remains a key challenge. To address this, we propose a novel PPG-to-ECG reconstruction method that leverages a Vision Transformer (ViT) as the core network. Unlike conventional approaches that rely on single-channel PPG, our method employs a four-channel signal image representation, incorporating the original PPG, its first-order difference, second-order difference, and area under the curve. This multi-channel design enriches feature extraction by preserving both temporal and physiological variations within the PPG. By leveraging the self-attention mechanism in ViT, our approach effectively captures both inter-beat and intra-beat dependencies, leading to more robust and accurate ECG reconstruction. Experimental results demonstrate that our method consistently outperforms existing 1D convolution-based approaches, achieving up to 29% reduction in PRD and 15% reduction in RMSE. The proposed approach also produces improvements in other evaluation metrics, highlighting its robustness and effectiveness in reconstructing ECG signals. Furthermore, to ensure a clinically relevant evaluation, we introduce new performance metrics, including QRS area error, PR interval error, RT interval error, and RT amplitude difference error. Our findings suggest that integrating a four-channel signal image representation with the self-attention mechanism of ViT enables more effective extraction of informative PPG features and improved modeling of beat-to-beat variations for PPG-to-ECG mapping. Beyond demonstrating the potential of PPG as a viable alternative for heart activity monitoring, our approach opens new avenues for cyclic signal analysis and prediction.
CLEP-GAN: An Innovative Approach to Subject-Independent ECG Reconstruction from PPG Signals
Feb 24, 2025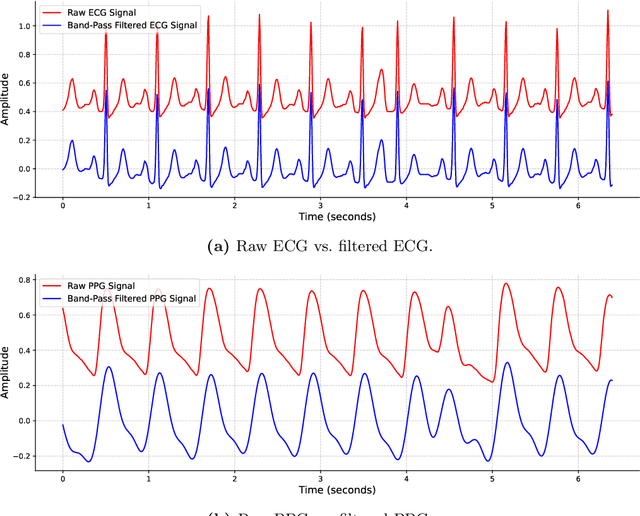
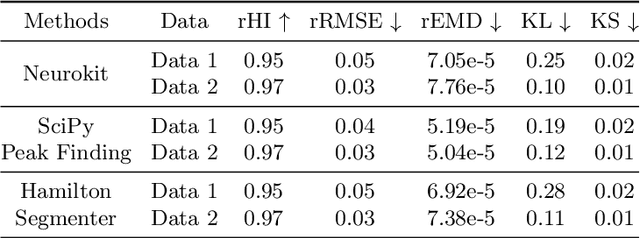
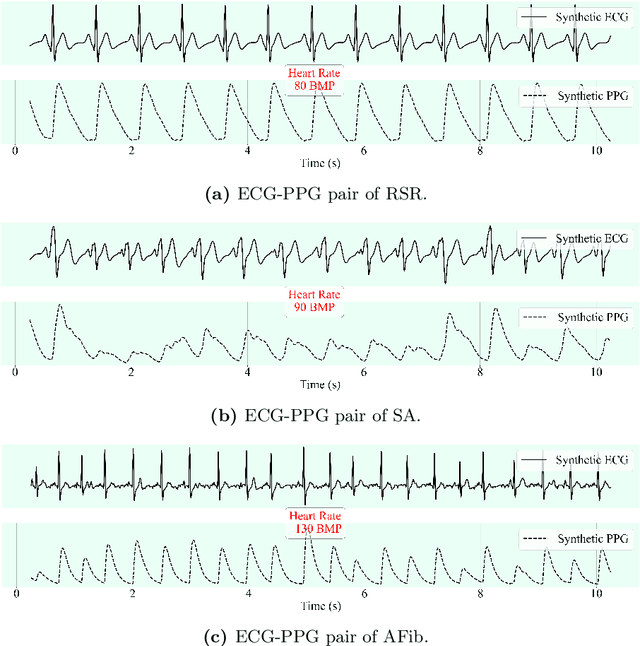
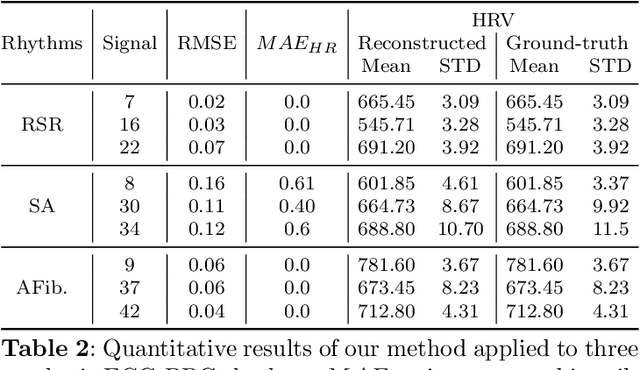
Abstract:This study addresses the challenge of reconstructing unseen ECG signals from PPG signals, a critical task for non-invasive cardiac monitoring. While numerous public ECG-PPG datasets are available, they lack the diversity seen in image datasets, and data collection processes often introduce noise, complicating ECG reconstruction from PPG even with advanced machine learning models. To tackle these challenges, we first introduce a novel synthetic ECG-PPG data generation technique using an ODE model to enhance training diversity. Next, we develop a novel subject-independent PPG-to-ECG reconstruction model that integrates contrastive learning, adversarial learning, and attention gating, achieving results comparable to or even surpassing existing approaches for unseen ECG reconstruction. Finally, we examine factors such as sex and age that impact reconstruction accuracy, emphasizing the importance of considering demographic diversity during model training and dataset augmentation.
FastOcc: Accelerating 3D Occupancy Prediction by Fusing the 2D Bird's-Eye View and Perspective View
Mar 05, 2024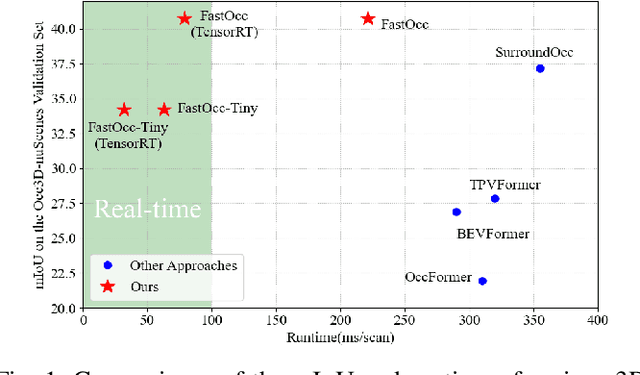
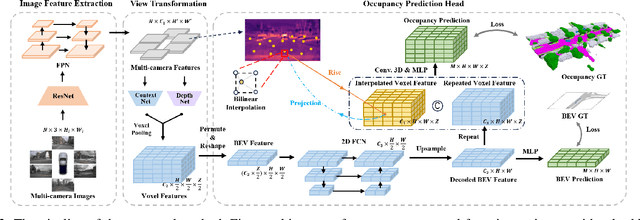
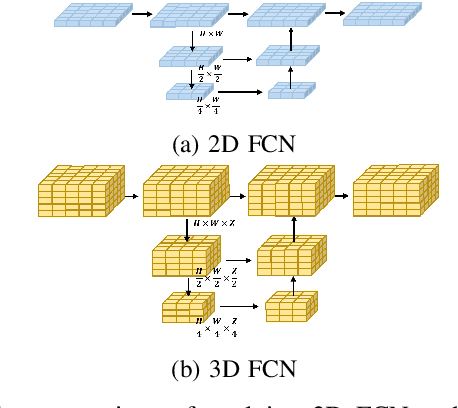
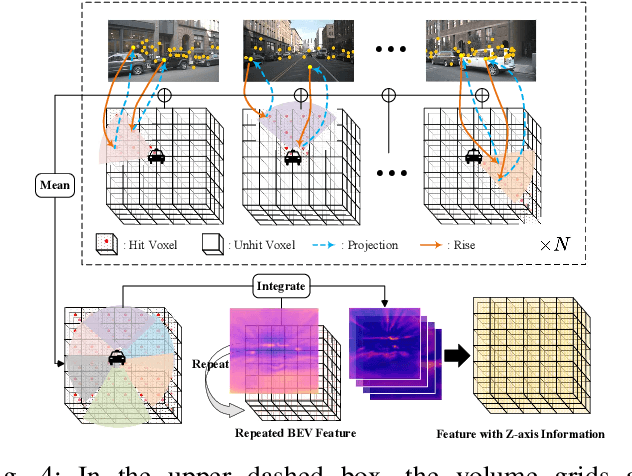
Abstract:In autonomous driving, 3D occupancy prediction outputs voxel-wise status and semantic labels for more comprehensive understandings of 3D scenes compared with traditional perception tasks, such as 3D object detection and bird's-eye view (BEV) semantic segmentation. Recent researchers have extensively explored various aspects of this task, including view transformation techniques, ground-truth label generation, and elaborate network design, aiming to achieve superior performance. However, the inference speed, crucial for running on an autonomous vehicle, is neglected. To this end, a new method, dubbed FastOcc, is proposed. By carefully analyzing the network effect and latency from four parts, including the input image resolution, image backbone, view transformation, and occupancy prediction head, it is found that the occupancy prediction head holds considerable potential for accelerating the model while keeping its accuracy. Targeted at improving this component, the time-consuming 3D convolution network is replaced with a novel residual-like architecture, where features are mainly digested by a lightweight 2D BEV convolution network and compensated by integrating the 3D voxel features interpolated from the original image features. Experiments on the Occ3D-nuScenes benchmark demonstrate that our FastOcc achieves state-of-the-art results with a fast inference speed.
OAG-Bench: A Human-Curated Benchmark for Academic Graph Mining
Feb 24, 2024



Abstract:With the rapid proliferation of scientific literature, versatile academic knowledge services increasingly rely on comprehensive academic graph mining. Despite the availability of public academic graphs, benchmarks, and datasets, these resources often fall short in multi-aspect and fine-grained annotations, are constrained to specific task types and domains, or lack underlying real academic graphs. In this paper, we present OAG-Bench, a comprehensive, multi-aspect, and fine-grained human-curated benchmark based on the Open Academic Graph (OAG). OAG-Bench covers 10 tasks, 20 datasets, 70+ baselines, and 120+ experimental results to date. We propose new data annotation strategies for certain tasks and offer a suite of data pre-processing codes, algorithm implementations, and standardized evaluation protocols to facilitate academic graph mining. Extensive experiments reveal that even advanced algorithms like large language models (LLMs) encounter difficulties in addressing key challenges in certain tasks, such as paper source tracing and scholar profiling. We also introduce the Open Academic Graph Challenge (OAG-Challenge) to encourage community input and sharing. We envisage that OAG-Bench can serve as a common ground for the community to evaluate and compare algorithms in academic graph mining, thereby accelerating algorithm development and advancement in this field. OAG-Bench is accessible at https://www.aminer.cn/data/.
Center Focusing Network for Real-Time LiDAR Panoptic Segmentation
Nov 16, 2023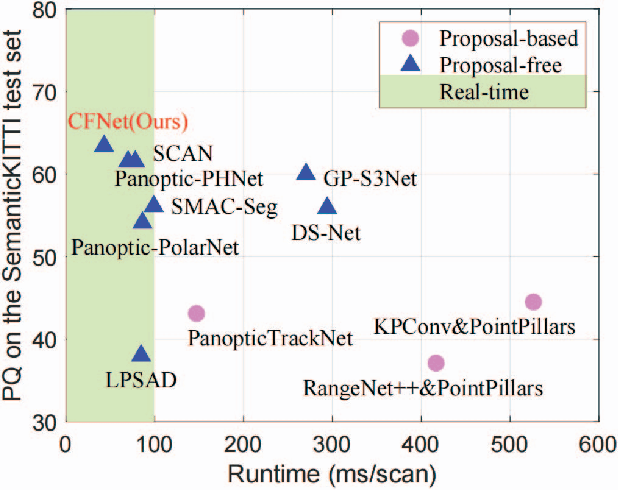
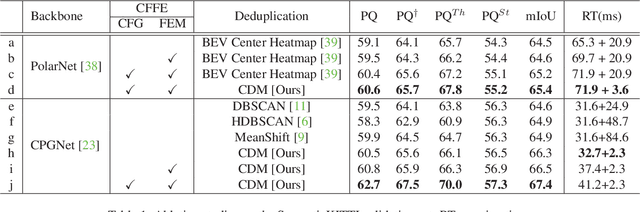
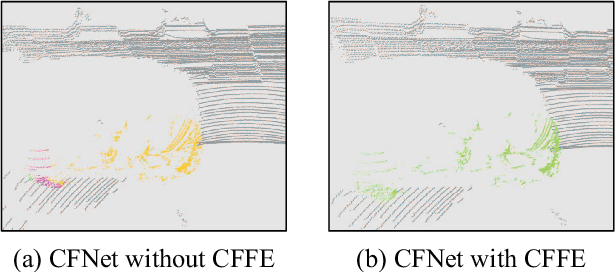
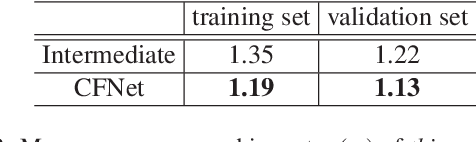
Abstract:LiDAR panoptic segmentation facilitates an autonomous vehicle to comprehensively understand the surrounding objects and scenes and is required to run in real time. The recent proposal-free methods accelerate the algorithm, but their effectiveness and efficiency are still limited owing to the difficulty of modeling non-existent instance centers and the costly center-based clustering modules. To achieve accurate and real-time LiDAR panoptic segmentation, a novel center focusing network (CFNet) is introduced. Specifically, the center focusing feature encoding (CFFE) is proposed to explicitly understand the relationships between the original LiDAR points and virtual instance centers by shifting the LiDAR points and filling in the center points. Moreover, to leverage the redundantly detected centers, a fast center deduplication module (CDM) is proposed to select only one center for each instance. Experiments on the SemanticKITTI and nuScenes panoptic segmentation benchmarks demonstrate that our CFNet outperforms all existing methods by a large margin and is 1.6 times faster than the most efficient method. The code is available at https://github.com/GangZhang842/CFNet.
Web-Scale Academic Name Disambiguation: the WhoIsWho Benchmark, Leaderboard, and Toolkit
Feb 23, 2023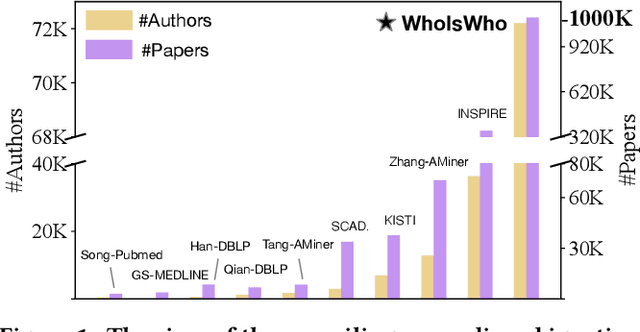

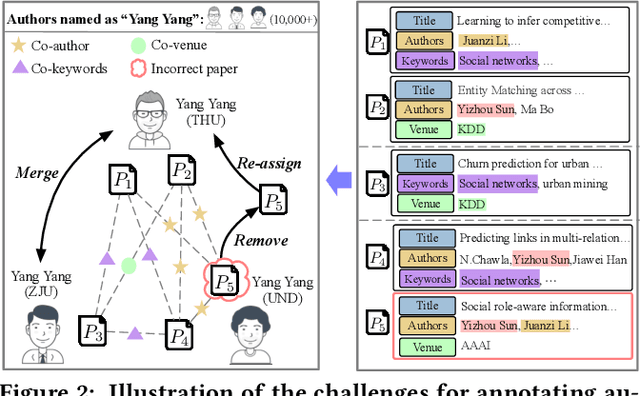
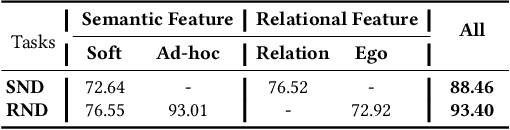
Abstract:Name disambiguation -- a fundamental problem in online academic systems -- is now facing greater challenges with the increasing growth of research papers. For example, on AMiner, an online academic search platform, about 10% of names own more than 100 authors. Such real-world hard cases cannot be fully addressed by existing research efforts, because of the small-scale or low-quality datasets that they use to build algorithms. The development of effective algorithms is further hampered by a variety of tasks and evaluation protocols designed on top of diverse datasets. To this end, we present WhoIsWho owning, a large-scale benchmark with over 1,000,000 papers built using an interactive annotation process, a regular leaderboard with comprehensive tasks, and an easy-to-use toolkit encapsulating the entire pipeline as well as the most powerful features and baseline models for tackling the tasks. Our developed strong baseline has already been deployed online in the AMiner system to enable daily arXiv paper assignments. The documentation and regular leaderboards are publicly available at http://whoiswho.biendata.xyz/.
EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks
Dec 06, 2022



Abstract:Background and Purpose: Colorectal cancer is a common fatal malignancy, the fourth most common cancer in men, and the third most common cancer in women worldwide. Timely detection of cancer in its early stages is essential for treating the disease. Currently, there is a lack of datasets for histopathological image segmentation of rectal cancer, which often hampers the assessment accuracy when computer technology is used to aid in diagnosis. Methods: This present study provided a new publicly available Enteroscope Biopsy Histopathological Hematoxylin and Eosin Image Dataset for Image Segmentation Tasks (EBHI-Seg). To demonstrate the validity and extensiveness of EBHI-Seg, the experimental results for EBHI-Seg are evaluated using classical machine learning methods and deep learning methods. Results: The experimental results showed that deep learning methods had a better image segmentation performance when utilizing EBHI-Seg. The maximum accuracy of the Dice evaluation metric for the classical machine learning method is 0.948, while the Dice evaluation metric for the deep learning method is 0.965. Conclusion: This publicly available dataset contained 5,170 images of six types of tumor differentiation stages and the corresponding ground truth images. The dataset can provide researchers with new segmentation algorithms for medical diagnosis of colorectal cancer, which can be used in the clinical setting to help doctors and patients.
An Algebraic Framework for Stock & Flow Diagrams and Dynamical Systems Using Category Theory
Nov 01, 2022



Abstract:Mathematical modeling of infectious disease at scale is important, but challenging. Some of these difficulties can be alleviated by an approach that takes diagrams seriously as mathematical formalisms in their own right. Stock & flow diagrams are widely used as broadly accessible building blocks for infectious disease modeling. In this chapter, rather than focusing on the underlying mathematics, we informally use communicable disease examples created by the implemented software of StockFlow.jl to explain the basics, characteristics, and benefits of the categorical framework. We first characterize categorical stock & flow diagrams, and note the clear separation between the syntax of stock & flow diagrams and their semantics, demonstrating three examples of semantics already implemented in the software: ODEs, causal loop diagrams, and system structure diagrams. We then establish composition and stratification frameworks and examples for stock & flow diagrams. Applying category theory, these frameworks can build large diagrams from smaller ones in a modular fashion. Finally, we introduce the open-source ModelCollab software for diagram-centric real-time collaborative modeling. Using the graphical user interface, this web-based software allows the user to undertake the types of categorically-rooted operations discussed above, but without any knowledge of their categorical foundations.
IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach
Jun 07, 2022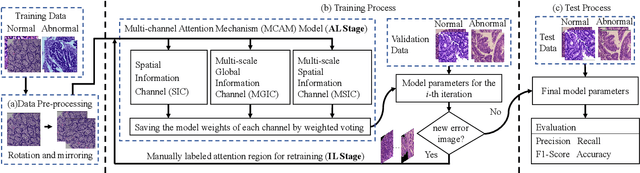
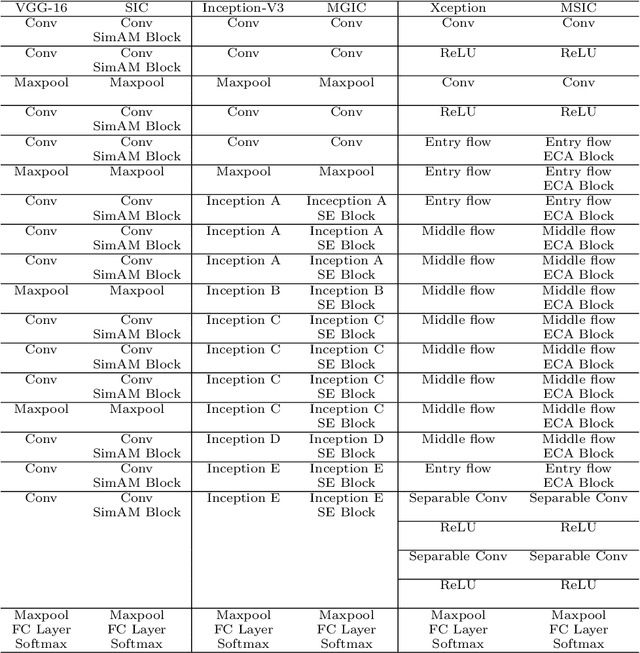
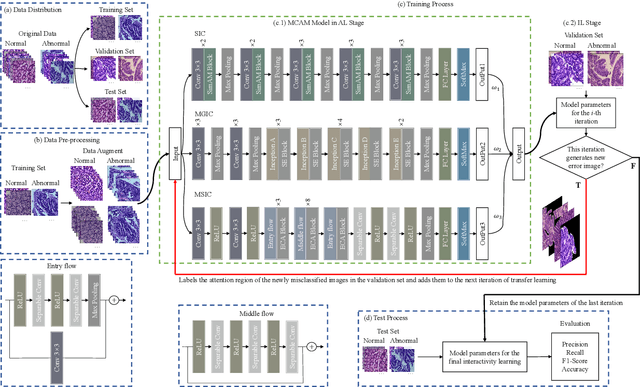
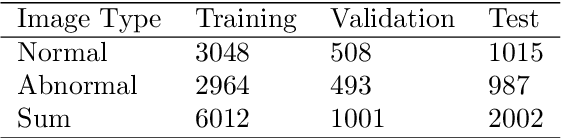
Abstract:In recent years, colorectal cancer has become one of the most significant diseases that endanger human health. Deep learning methods are increasingly important for the classification of colorectal histopathology images. However, existing approaches focus more on end-to-end automatic classification using computers rather than human-computer interaction. In this paper, we propose an IL-MCAM framework. It is based on attention mechanisms and interactive learning. The proposed IL-MCAM framework includes two stages: automatic learning (AL) and interactivity learning (IL). In the AL stage, a multi-channel attention mechanism model containing three different attention mechanism channels and convolutional neural networks is used to extract multi-channel features for classification. In the IL stage, the proposed IL-MCAM framework continuously adds misclassified images to the training set in an interactive approach, which improves the classification ability of the MCAM model. We carried out a comparison experiment on our dataset and an extended experiment on the HE-NCT-CRC-100K dataset to verify the performance of the proposed IL-MCAM framework, achieving classification accuracies of 98.98% and 99.77%, respectively. In addition, we conducted an ablation experiment and an interchangeability experiment to verify the ability and interchangeability of the three channels. The experimental results show that the proposed IL-MCAM framework has excellent performance in the colorectal histopathological image classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge