Md Mamunur Rahaman
Leveraging Vision-Language Embeddings for Zero-Shot Learning in Histopathology Images
Mar 13, 2025Abstract:Zero-shot learning holds tremendous potential for histopathology image analysis by enabling models to generalize to unseen classes without extensive labeled data. Recent advancements in vision-language models (VLMs) have expanded the capabilities of ZSL, allowing models to perform tasks without task-specific fine-tuning. However, applying VLMs to histopathology presents considerable challenges due to the complexity of histopathological imagery and the nuanced nature of diagnostic tasks. In this paper, we propose a novel framework called Multi-Resolution Prompt-guided Hybrid Embedding (MR-PHE) to address these challenges in zero-shot histopathology image classification. MR-PHE leverages multiresolution patch extraction to mimic the diagnostic workflow of pathologists, capturing both fine-grained cellular details and broader tissue structures critical for accurate diagnosis. We introduce a hybrid embedding strategy that integrates global image embeddings with weighted patch embeddings, effectively combining local and global contextual information. Additionally, we develop a comprehensive prompt generation and selection framework, enriching class descriptions with domain-specific synonyms and clinically relevant features to enhance semantic understanding. A similarity-based patch weighting mechanism assigns attention-like weights to patches based on their relevance to class embeddings, emphasizing diagnostically important regions during classification. Our approach utilizes pretrained VLM, CONCH for ZSL without requiring domain-specific fine-tuning, offering scalability and reducing dependence on large annotated datasets. Experimental results demonstrate that MR-PHE not only significantly improves zero-shot classification performance on histopathology datasets but also often surpasses fully supervised models.
Breast Cancer Histopathology Image based Gene Expression Prediction using Spatial Transcriptomics data and Deep Learning
Mar 17, 2023Abstract:Tumour heterogeneity in breast cancer poses challenges in predicting outcome and response to therapy. Spatial transcriptomics technologies may address these challenges, as they provide a wealth of information about gene expression at the cell level, but they are expensive, hindering their use in large-scale clinical oncology studies. Predicting gene expression from hematoxylin and eosin stained histology images provides a more affordable alternative for such studies. Here we present BrST-Net, a deep learning framework for predicting gene expression from histopathology images using spatial transcriptomics data. Using this framework, we trained and evaluated 10 state-of-the-art deep learning models without utilizing pretrained weights for the prediction of 250 genes. To enhance the generalisation performance of the main network, we introduce an auxiliary network into the framework. Our methodology outperforms previous studies, with 237 genes identified with positive correlation, including 24 genes with a median correlation coefficient greater than 0.50. This is a notable improvement over previous studies, which could predict only 102 genes with positive correlation, with the highest correlation values ranging from 0.29 to 0.34.
ACTIVE: A Deep Model for Sperm and Impurity Detection in Microscopic Videos
Jan 15, 2023



Abstract:The accurate detection of sperms and impurities is a very challenging task, facing problems such as the small size of targets, indefinite target morphologies, low contrast and resolution of the video, and similarity of sperms and impurities. So far, the detection of sperms and impurities still largely relies on the traditional image processing and detection techniques which only yield limited performance and often require manual intervention in the detection process, therefore unfavorably escalating the time cost and injecting the subjective bias into the analysis. Encouraged by the successes of deep learning methods in numerous object detection tasks, here we report a deep learning model based on Double Branch Feature Extraction Network (DBFEN) and Cross-conjugate Feature Pyramid Networks (CCFPN).DBFEN is designed to extract visual features from tiny objects with a double branch structure, and CCFPN is further introduced to fuse the features extracted by DBFEN to enhance the description of position and high-level semantic information. Our work is the pioneer of introducing deep learning approaches to the detection of sperms and impurities. Experiments show that the highest AP50 of the sperm and impurity detection is 91.13% and 59.64%, which lead its competitors by a substantial margin and establish new state-of-the-art results in this problem.
Segmentation of Weakly Visible Environmental Microorganism Images Using Pair-wise Deep Learning Features
Aug 31, 2022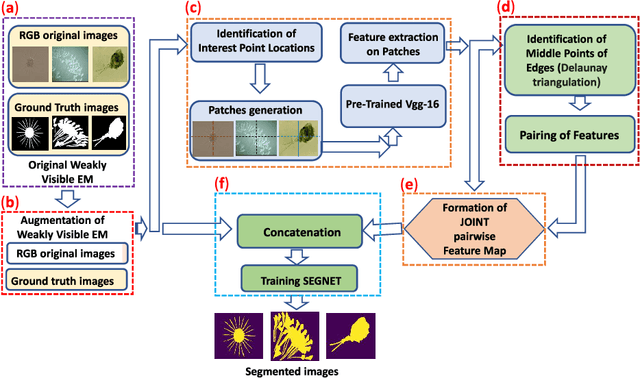



Abstract:The use of Environmental Microorganisms (EMs) offers a highly efficient, low cost and harmless remedy to environmental pollution, by monitoring and decomposing of pollutants. This relies on how the EMs are correctly segmented and identified. With the aim of enhancing the segmentation of weakly visible EM images which are transparent, noisy and have low contrast, a Pairwise Deep Learning Feature Network (PDLF-Net) is proposed in this study. The use of PDLFs enables the network to focus more on the foreground (EMs) by concatenating the pairwise deep learning features of each image to different blocks of the base model SegNet. Leveraging the Shi and Tomas descriptors, we extract each image's deep features on the patches, which are centered at each descriptor using the VGG-16 model. Then, to learn the intermediate characteristics between the descriptors, pairing of the features is performed based on the Delaunay triangulation theorem to form pairwise deep learning features. In this experiment, the PDLF-Net achieves outstanding segmentation results of 89.24%, 63.20%, 77.27%, 35.15%, 89.72%, 91.44% and 89.30% on the accuracy, IoU, Dice, VOE, sensitivity, precision and specificity, respectively.
IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach
Jun 07, 2022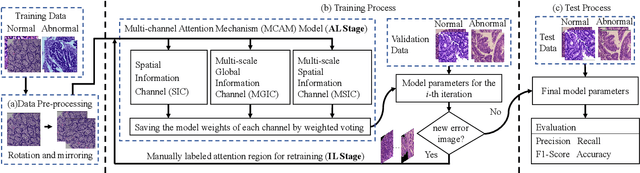
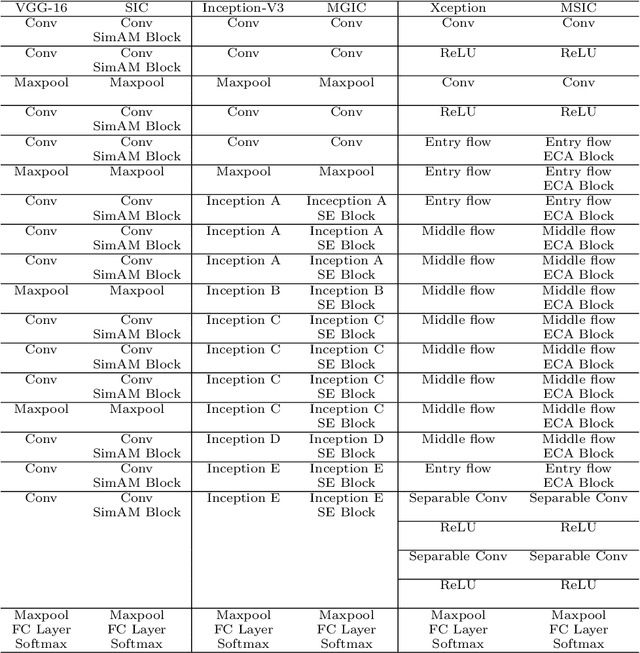
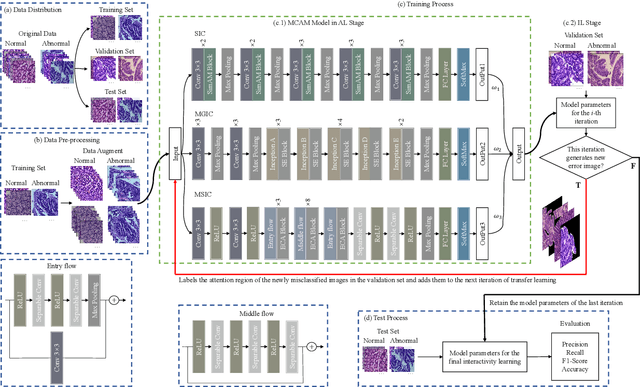
Abstract:In recent years, colorectal cancer has become one of the most significant diseases that endanger human health. Deep learning methods are increasingly important for the classification of colorectal histopathology images. However, existing approaches focus more on end-to-end automatic classification using computers rather than human-computer interaction. In this paper, we propose an IL-MCAM framework. It is based on attention mechanisms and interactive learning. The proposed IL-MCAM framework includes two stages: automatic learning (AL) and interactivity learning (IL). In the AL stage, a multi-channel attention mechanism model containing three different attention mechanism channels and convolutional neural networks is used to extract multi-channel features for classification. In the IL stage, the proposed IL-MCAM framework continuously adds misclassified images to the training set in an interactive approach, which improves the classification ability of the MCAM model. We carried out a comparison experiment on our dataset and an extended experiment on the HE-NCT-CRC-100K dataset to verify the performance of the proposed IL-MCAM framework, achieving classification accuracies of 98.98% and 99.77%, respectively. In addition, we conducted an ablation experiment and an interchangeability experiment to verify the ability and interchangeability of the three channels. The experimental results show that the proposed IL-MCAM framework has excellent performance in the colorectal histopathological image classification tasks.
CVM-Cervix: A Hybrid Cervical Pap-Smear Image Classification Framework Using CNN, Visual Transformer and Multilayer Perceptron
Jun 02, 2022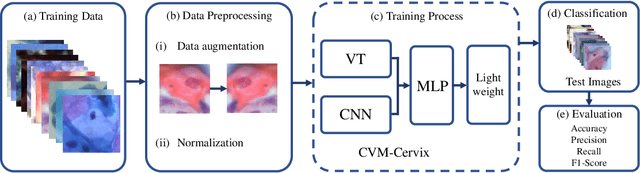

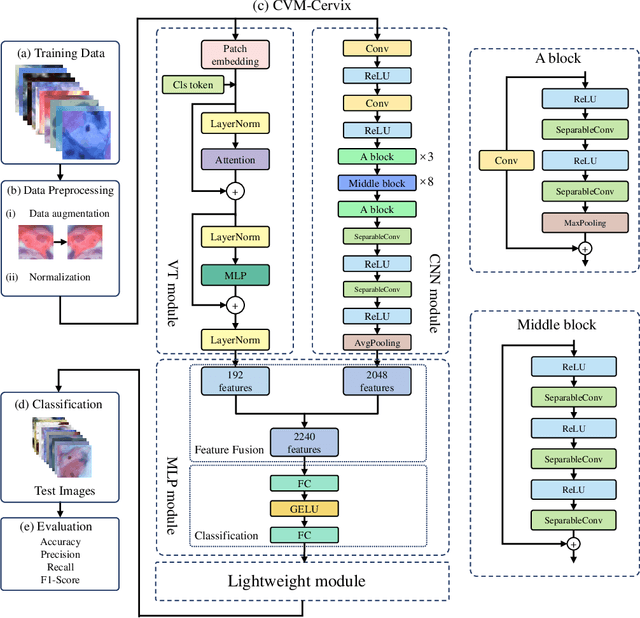

Abstract:Cervical cancer is the seventh most common cancer among all the cancers worldwide and the fourth most common cancer among women. Cervical cytopathology image classification is an important method to diagnose cervical cancer. Manual screening of cytopathology images is time-consuming and error-prone. The emergence of the automatic computer-aided diagnosis system solves this problem. This paper proposes a framework called CVM-Cervix based on deep learning to perform cervical cell classification tasks. It can analyze pap slides quickly and accurately. CVM-Cervix first proposes a Convolutional Neural Network module and a Visual Transformer module for local and global feature extraction respectively, then a Multilayer Perceptron module is designed to fuse the local and global features for the final classification. Experimental results show the effectiveness and potential of the proposed CVM-Cervix in the field of cervical Pap smear image classification. In addition, according to the practical needs of clinical work, we perform a lightweight post-processing to compress the model.
An application of Pixel Interval Down-sampling (PID) for dense tiny microorganism counting on environmental microorganism images
Apr 04, 2022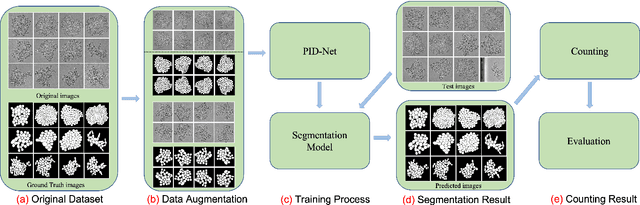
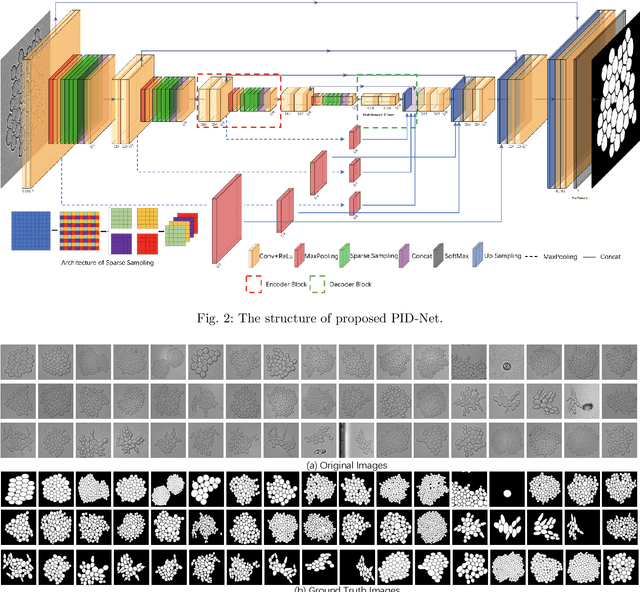
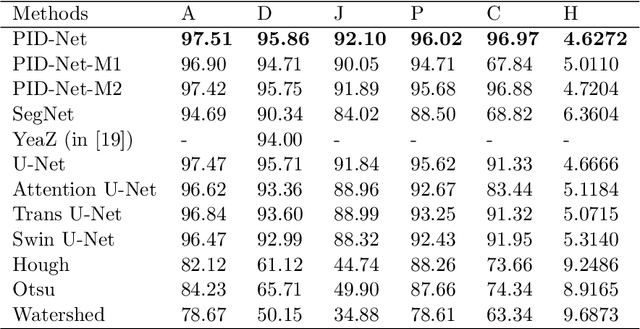
Abstract:This paper proposes a novel pixel interval down-sampling network (PID-Net) for dense tiny objects (yeast cells) counting tasks with higher accuracy. The PID-Net is an end-to-end CNN model with encoder to decoder architecture. The pixel interval down-sampling operations are concatenated with max-pooling operations to combine the sparse and dense features. It addresses the limitation of contour conglutination of dense objects while counting. Evaluation was done using classical segmentation metrics (Dice, Jaccard, Hausdorff distance) as well as counting metrics. Experimental result shows that the proposed PID-Net has the best performance and potential for dense tiny objects counting tasks, which achieves 96.97% counting accuracy on the dataset with 2448 yeast cell images. By comparing with the state-of-the-art approaches like Attention U-Net, Swin U-Net and Trans U-Net, the proposed PID-Net can segment the dense tiny objects with clearer boundaries and fewer incorrect debris, which shows the great potential of PID-Net in the task of accurate counting tasks.
A Comprehensive Survey with Quantitative Comparison of Image Analysis Methods for Microorganism Biovolume Measurements
Feb 18, 2022
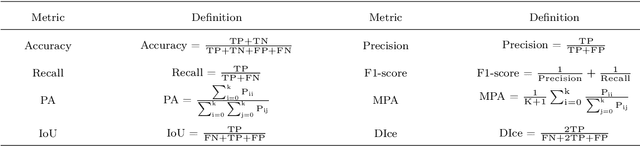
Abstract:With the acceleration of urbanization and living standards, microorganisms play increasingly important roles in industrial production, bio-technique, and food safety testing. Microorganism biovolume measurements are one of the essential parts of microbial analysis. However, traditional manual measurement methods are time-consuming and challenging to measure the characteristics precisely. With the development of digital image processing techniques, the characteristics of the microbial population can be detected and quantified. The changing trend can be adjusted in time and provided a basis for the improvement. The applications of the microorganism biovolume measurement method have developed since the 1980s. More than 60 articles are reviewed in this study, and the articles are grouped by digital image segmentation methods with periods. This study has high research significance and application value, which can be referred to microbial researchers to have a comprehensive understanding of microorganism biovolume measurements using digital image analysis methods and potential applications.
EBHI:A New Enteroscope Biopsy Histopathological H&E Image Dataset for Image Classification Evaluation
Feb 17, 2022
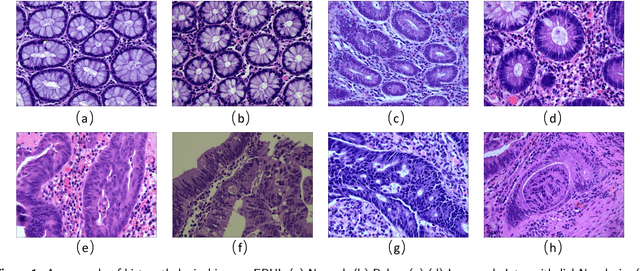
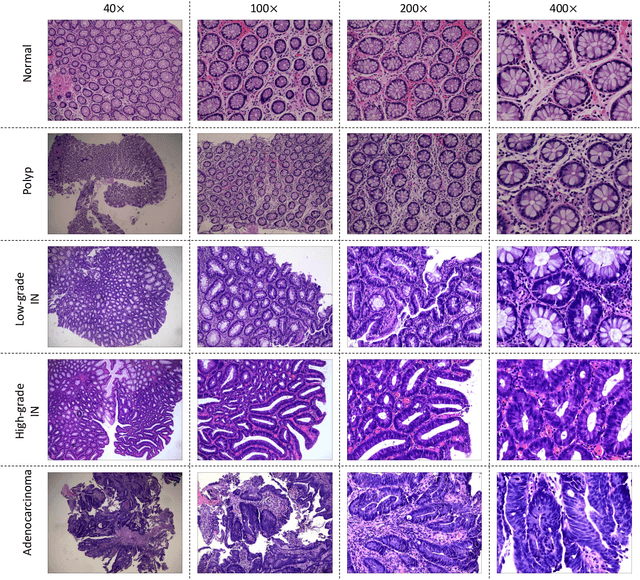
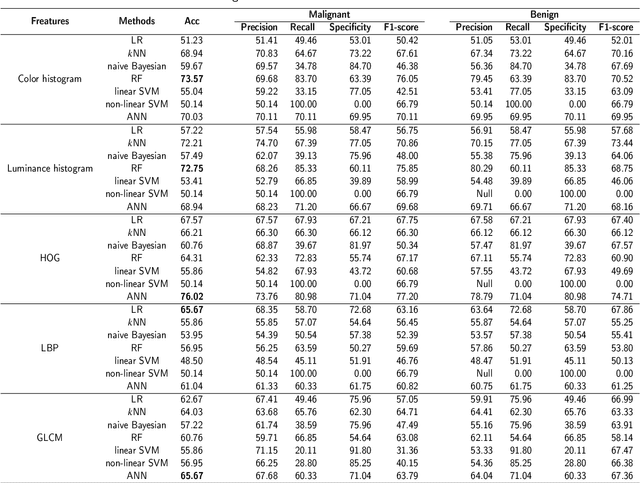
Abstract:Background and purpose: Colorectal cancer has become the third most common cancer worldwide, accounting for approximately 10% of cancer patients. Early detection of the disease is important for the treatment of colorectal cancer patients. Histopathological examination is the gold standard for screening colorectal cancer. However, the current lack of histopathological image datasets of colorectal cancer, especially enteroscope biopsies, hinders the accurate evaluation of computer-aided diagnosis techniques. Methods: A new publicly available Enteroscope Biopsy Histopathological H&E Image Dataset (EBHI) is published in this paper. To demonstrate the effectiveness of the EBHI dataset, we have utilized several machine learning, convolutional neural networks and novel transformer-based classifiers for experimentation and evaluation, using an image with a magnification of 200x. Results: Experimental results show that the deep learning method performs well on the EBHI dataset. Traditional machine learning methods achieve maximum accuracy of 76.02% and deep learning method achieves a maximum accuracy of 95.37%. Conclusion: To the best of our knowledge, EBHI is the first publicly available colorectal histopathology enteroscope biopsy dataset with four magnifications and five types of images of tumor differentiation stages, totaling 5532 images. We believe that EBHI could attract researchers to explore new classification algorithms for the automated diagnosis of colorectal cancer, which could help physicians and patients in clinical settings.
What Can Machine Vision Do for Lymphatic Histopathology Image Analysis: A Comprehensive Review
Jan 21, 2022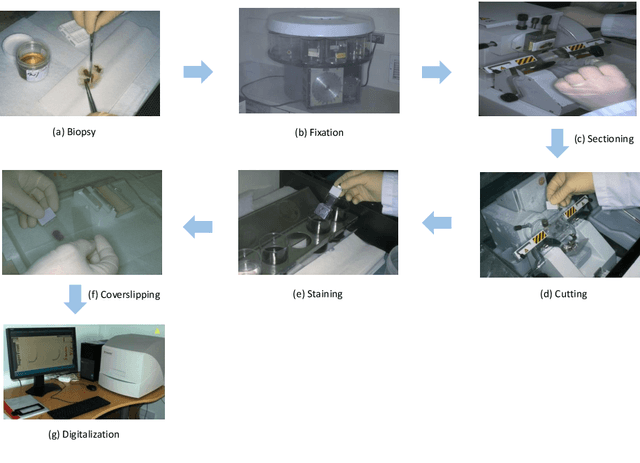
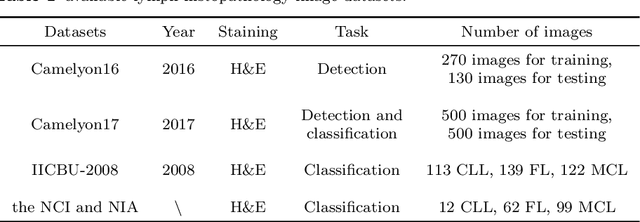
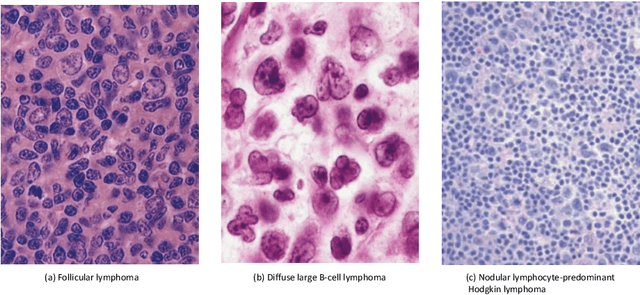
Abstract:In the past ten years, the computing power of machine vision (MV) has been continuously improved, and image analysis algorithms have developed rapidly. At the same time, histopathological slices can be stored as digital images. Therefore, MV algorithms can provide doctors with diagnostic references. In particular, the continuous improvement of deep learning algorithms has further improved the accuracy of MV in disease detection and diagnosis. This paper reviews the applications of image processing technology based on MV in lymphoma histopathological images in recent years, including segmentation, classification and detection. Finally, the current methods are analyzed, some more potential methods are proposed, and further prospects are made.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge