Hongzan Sun
ECPC-IDS:A benchmark endometrail cancer PET/CT image dataset for evaluation of semantic segmentation and detection of hypermetabolic regions
Sep 02, 2023



Abstract:Endometrial cancer is one of the most common tumors in the female reproductive system and is the third most common gynecological malignancy that causes death after ovarian and cervical cancer. Early diagnosis can significantly improve the 5-year survival rate of patients. With the development of artificial intelligence, computer-assisted diagnosis plays an increasingly important role in improving the accuracy and objectivity of diagnosis, as well as reducing the workload of doctors. However, the absence of publicly available endometrial cancer image datasets restricts the application of computer-assisted diagnostic techniques.In this paper, a publicly available Endometrial Cancer PET/CT Image Dataset for Evaluation of Semantic Segmentation and Detection of Hypermetabolic Regions (ECPC-IDS) are published. Specifically, the segmentation section includes PET and CT images, with a total of 7159 images in multiple formats. In order to prove the effectiveness of segmentation methods on ECPC-IDS, five classical deep learning semantic segmentation methods are selected to test the image segmentation task. The object detection section also includes PET and CT images, with a total of 3579 images and XML files with annotation information. Six deep learning methods are selected for experiments on the detection task.This study conduct extensive experiments using deep learning-based semantic segmentation and object detection methods to demonstrate the differences between various methods on ECPC-IDS. As far as we know, this is the first publicly available dataset of endometrial cancer with a large number of multiple images, including a large amount of information required for image and target detection. ECPC-IDS can aid researchers in exploring new algorithms to enhance computer-assisted technology, benefiting both clinical doctors and patients greatly.
PHE-SICH-CT-IDS: A Benchmark CT Image Dataset for Evaluation Semantic Segmentation, Object Detection and Radiomic Feature Extraction of Perihematomal Edema in Spontaneous Intracerebral Hemorrhage
Aug 21, 2023Abstract:Intracerebral hemorrhage is one of the diseases with the highest mortality and poorest prognosis worldwide. Spontaneous intracerebral hemorrhage (SICH) typically presents acutely, prompt and expedited radiological examination is crucial for diagnosis, localization, and quantification of the hemorrhage. Early detection and accurate segmentation of perihematomal edema (PHE) play a critical role in guiding appropriate clinical intervention and enhancing patient prognosis. However, the progress and assessment of computer-aided diagnostic methods for PHE segmentation and detection face challenges due to the scarcity of publicly accessible brain CT image datasets. This study establishes a publicly available CT dataset named PHE-SICH-CT-IDS for perihematomal edema in spontaneous intracerebral hemorrhage. The dataset comprises 120 brain CT scans and 7,022 CT images, along with corresponding medical information of the patients. To demonstrate its effectiveness, classical algorithms for semantic segmentation, object detection, and radiomic feature extraction are evaluated. The experimental results confirm the suitability of PHE-SICH-CT-IDS for assessing the performance of segmentation, detection and radiomic feature extraction methods. To the best of our knowledge, this is the first publicly available dataset for PHE in SICH, comprising various data formats suitable for applications across diverse medical scenarios. We believe that PHE-SICH-CT-IDS will allure researchers to explore novel algorithms, providing valuable support for clinicians and patients in the clinical setting. PHE-SICH-CT-IDS is freely published for non-commercial purpose at: https://figshare.com/articles/dataset/PHE-SICH-CT-IDS/23957937.
AATCT-IDS: A Benchmark Abdominal Adipose Tissue CT Image Dataset for Image Denoising, Semantic Segmentation, and Radiomics Evaluation
Aug 16, 2023Abstract:Methods: In this study, a benchmark \emph{Abdominal Adipose Tissue CT Image Dataset} (AATTCT-IDS) containing 300 subjects is prepared and published. AATTCT-IDS publics 13,732 raw CT slices, and the researchers individually annotate the subcutaneous and visceral adipose tissue regions of 3,213 of those slices that have the same slice distance to validate denoising methods, train semantic segmentation models, and study radiomics. For different tasks, this paper compares and analyzes the performance of various methods on AATTCT-IDS by combining the visualization results and evaluation data. Thus, verify the research potential of this data set in the above three types of tasks. Results: In the comparative study of image denoising, algorithms using a smoothing strategy suppress mixed noise at the expense of image details and obtain better evaluation data. Methods such as BM3D preserve the original image structure better, although the evaluation data are slightly lower. The results show significant differences among them. In the comparative study of semantic segmentation of abdominal adipose tissue, the segmentation results of adipose tissue by each model show different structural characteristics. Among them, BiSeNet obtains segmentation results only slightly inferior to U-Net with the shortest training time and effectively separates small and isolated adipose tissue. In addition, the radiomics study based on AATTCT-IDS reveals three adipose distributions in the subject population. Conclusion: AATTCT-IDS contains the ground truth of adipose tissue regions in abdominal CT slices. This open-source dataset can attract researchers to explore the multi-dimensional characteristics of abdominal adipose tissue and thus help physicians and patients in clinical practice. AATCT-IDS is freely published for non-commercial purpose at: \url{https://figshare.com/articles/dataset/AATTCT-IDS/23807256}.
ACTIVE: A Deep Model for Sperm and Impurity Detection in Microscopic Videos
Jan 15, 2023



Abstract:The accurate detection of sperms and impurities is a very challenging task, facing problems such as the small size of targets, indefinite target morphologies, low contrast and resolution of the video, and similarity of sperms and impurities. So far, the detection of sperms and impurities still largely relies on the traditional image processing and detection techniques which only yield limited performance and often require manual intervention in the detection process, therefore unfavorably escalating the time cost and injecting the subjective bias into the analysis. Encouraged by the successes of deep learning methods in numerous object detection tasks, here we report a deep learning model based on Double Branch Feature Extraction Network (DBFEN) and Cross-conjugate Feature Pyramid Networks (CCFPN).DBFEN is designed to extract visual features from tiny objects with a double branch structure, and CCFPN is further introduced to fuse the features extracted by DBFEN to enhance the description of position and high-level semantic information. Our work is the pioneer of introducing deep learning approaches to the detection of sperms and impurities. Experiments show that the highest AP50 of the sperm and impurity detection is 91.13% and 59.64%, which lead its competitors by a substantial margin and establish new state-of-the-art results in this problem.
EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks
Dec 06, 2022



Abstract:Background and Purpose: Colorectal cancer is a common fatal malignancy, the fourth most common cancer in men, and the third most common cancer in women worldwide. Timely detection of cancer in its early stages is essential for treating the disease. Currently, there is a lack of datasets for histopathological image segmentation of rectal cancer, which often hampers the assessment accuracy when computer technology is used to aid in diagnosis. Methods: This present study provided a new publicly available Enteroscope Biopsy Histopathological Hematoxylin and Eosin Image Dataset for Image Segmentation Tasks (EBHI-Seg). To demonstrate the validity and extensiveness of EBHI-Seg, the experimental results for EBHI-Seg are evaluated using classical machine learning methods and deep learning methods. Results: The experimental results showed that deep learning methods had a better image segmentation performance when utilizing EBHI-Seg. The maximum accuracy of the Dice evaluation metric for the classical machine learning method is 0.948, while the Dice evaluation metric for the deep learning method is 0.965. Conclusion: This publicly available dataset contained 5,170 images of six types of tumor differentiation stages and the corresponding ground truth images. The dataset can provide researchers with new segmentation algorithms for medical diagnosis of colorectal cancer, which can be used in the clinical setting to help doctors and patients.
IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach
Jun 07, 2022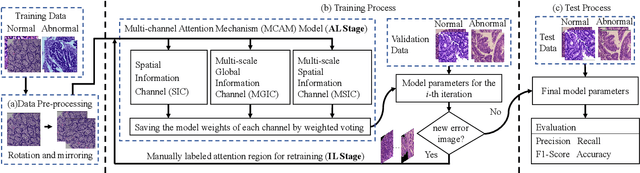
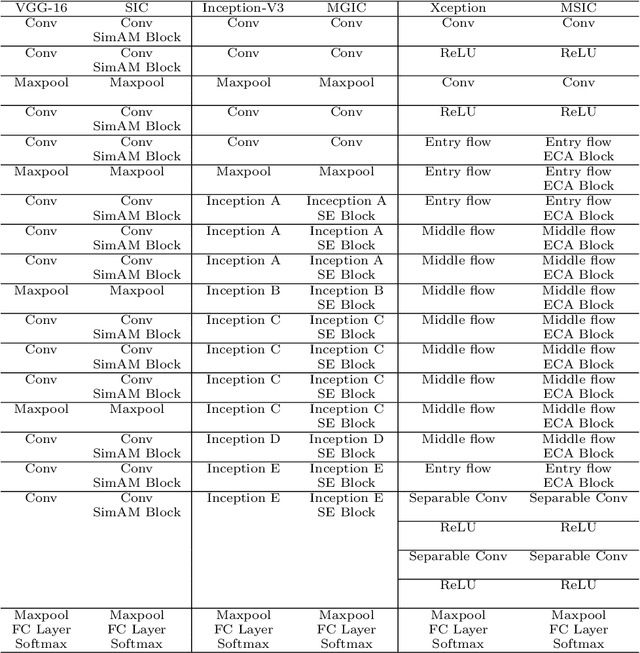
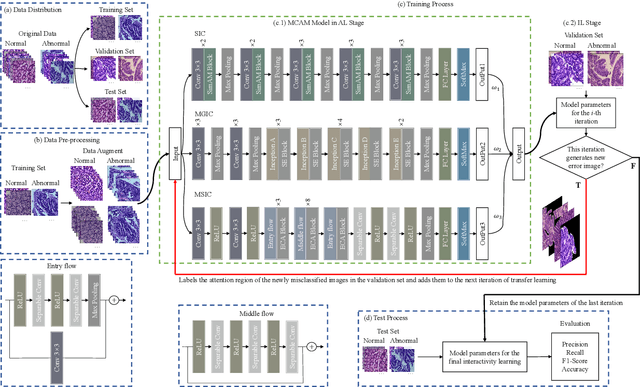
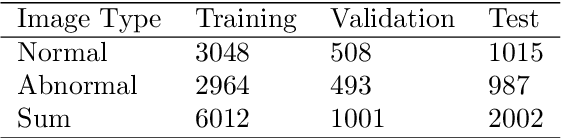
Abstract:In recent years, colorectal cancer has become one of the most significant diseases that endanger human health. Deep learning methods are increasingly important for the classification of colorectal histopathology images. However, existing approaches focus more on end-to-end automatic classification using computers rather than human-computer interaction. In this paper, we propose an IL-MCAM framework. It is based on attention mechanisms and interactive learning. The proposed IL-MCAM framework includes two stages: automatic learning (AL) and interactivity learning (IL). In the AL stage, a multi-channel attention mechanism model containing three different attention mechanism channels and convolutional neural networks is used to extract multi-channel features for classification. In the IL stage, the proposed IL-MCAM framework continuously adds misclassified images to the training set in an interactive approach, which improves the classification ability of the MCAM model. We carried out a comparison experiment on our dataset and an extended experiment on the HE-NCT-CRC-100K dataset to verify the performance of the proposed IL-MCAM framework, achieving classification accuracies of 98.98% and 99.77%, respectively. In addition, we conducted an ablation experiment and an interchangeability experiment to verify the ability and interchangeability of the three channels. The experimental results show that the proposed IL-MCAM framework has excellent performance in the colorectal histopathological image classification tasks.
CVM-Cervix: A Hybrid Cervical Pap-Smear Image Classification Framework Using CNN, Visual Transformer and Multilayer Perceptron
Jun 02, 2022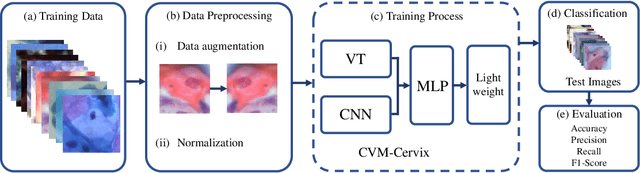

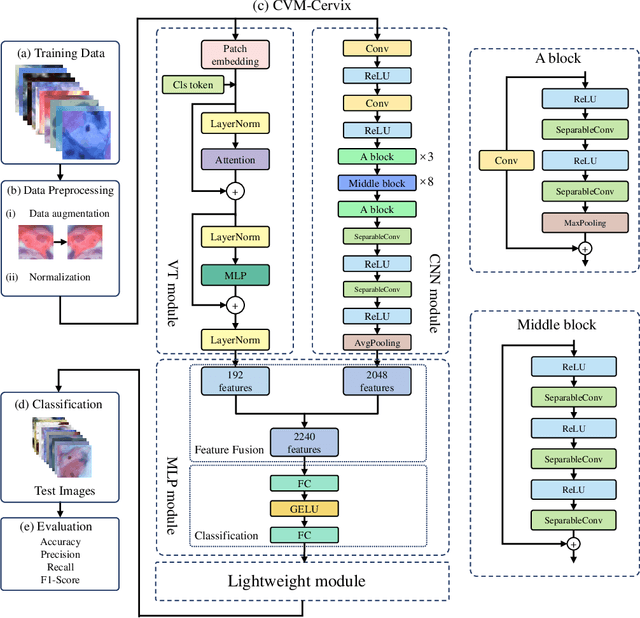

Abstract:Cervical cancer is the seventh most common cancer among all the cancers worldwide and the fourth most common cancer among women. Cervical cytopathology image classification is an important method to diagnose cervical cancer. Manual screening of cytopathology images is time-consuming and error-prone. The emergence of the automatic computer-aided diagnosis system solves this problem. This paper proposes a framework called CVM-Cervix based on deep learning to perform cervical cell classification tasks. It can analyze pap slides quickly and accurately. CVM-Cervix first proposes a Convolutional Neural Network module and a Visual Transformer module for local and global feature extraction respectively, then a Multilayer Perceptron module is designed to fuse the local and global features for the final classification. Experimental results show the effectiveness and potential of the proposed CVM-Cervix in the field of cervical Pap smear image classification. In addition, according to the practical needs of clinical work, we perform a lightweight post-processing to compress the model.
A Comparative Study of Gastric Histopathology Sub-size Image Classification: from Linear Regression to Visual Transformer
May 25, 2022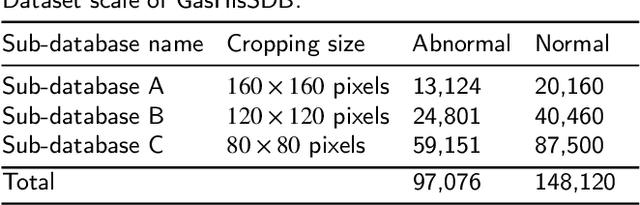
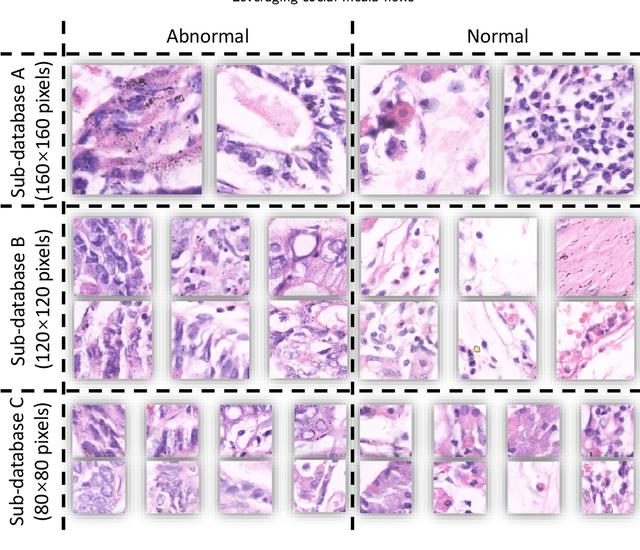
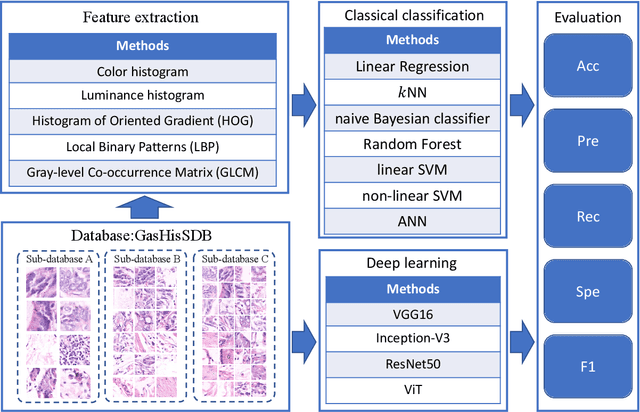
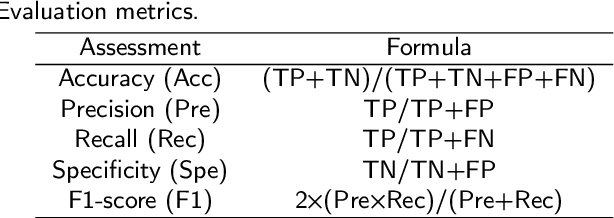
Abstract:Gastric cancer is the fifth most common cancer in the world. At the same time, it is also the fourth most deadly cancer. Early detection of cancer exists as a guide for the treatment of gastric cancer. Nowadays, computer technology has advanced rapidly to assist physicians in the diagnosis of pathological pictures of gastric cancer. Ensemble learning is a way to improve the accuracy of algorithms, and finding multiple learning models with complementarity types is the basis of ensemble learning. The complementarity of sub-size pathology image classifiers when machine performance is insufficient is explored in this experimental platform. We choose seven classical machine learning classifiers and four deep learning classifiers for classification experiments on the GasHisSDB database. Among them, classical machine learning algorithms extract five different image virtual features to match multiple classifier algorithms. For deep learning, we choose three convolutional neural network classifiers. In addition, we also choose a novel Transformer-based classifier. The experimental platform, in which a large number of classical machine learning and deep learning methods are performed, demonstrates that there are differences in the performance of different classifiers on GasHisSDB. Classical machine learning models exist for classifiers that classify Abnormal categories very well, while classifiers that excel in classifying Normal categories also exist. Deep learning models also exist with multiple models that can be complementarity. Suitable classifiers are selected for ensemble learning, when machine performance is insufficient. This experimental platform demonstrates that multiple classifiers are indeed complementarity and can improve the efficiency of ensemble learning. This can better assist doctors in diagnosis, improve the detection of gastric cancer, and increase the cure rate.
Application of Graph Based Features in Computer Aided Diagnosis for Histopathological Image Classification of Gastric Cancer
May 17, 2022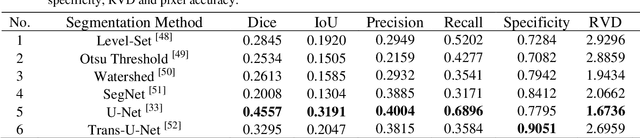
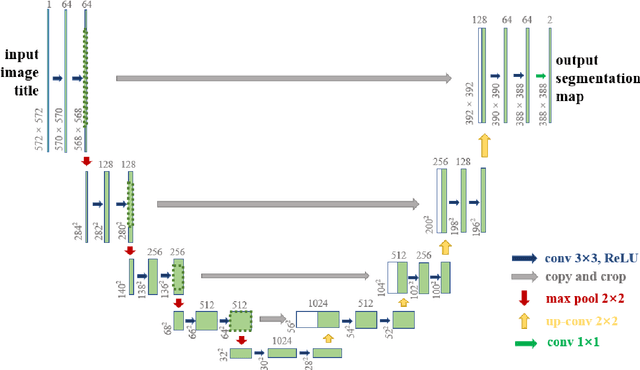

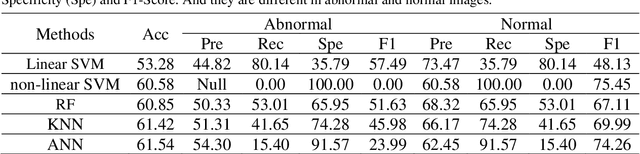
Abstract:The gold standard for gastric cancer detection is gastric histopathological image analysis, but there are certain drawbacks in the existing histopathological detection and diagnosis. In this paper, based on the study of computer aided diagnosis system, graph based features are applied to gastric cancer histopathology microscopic image analysis, and a classifier is used to classify gastric cancer cells from benign cells. Firstly, image segmentation is performed, and after finding the region, cell nuclei are extracted using the k-means method, the minimum spanning tree (MST) is drawn, and graph based features of the MST are extracted. The graph based features are then put into the classifier for classification. In this study, different segmentation methods are compared in the tissue segmentation stage, among which are Level-Set, Otsu thresholding, watershed, SegNet, U-Net and Trans-U-Net segmentation; Graph based features, Red, Green, Blue features, Grey-Level Co-occurrence Matrix features, Histograms of Oriented Gradient features and Local Binary Patterns features are compared in the feature extraction stage; Radial Basis Function (RBF) Support Vector Machine (SVM), Linear SVM, Artificial Neural Network, Random Forests, k-NearestNeighbor, VGG16, and Inception-V3 are compared in the classifier stage. It is found that using U-Net to segment tissue areas, then extracting graph based features, and finally using RBF SVM classifier gives the optimal results with 94.29%.
Application of Transfer Learning and Ensemble Learning in Image-level Classification for Breast Histopathology
Apr 18, 2022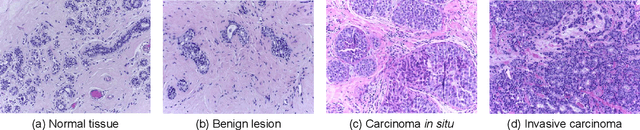
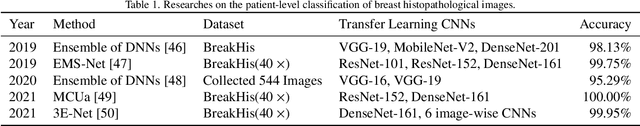
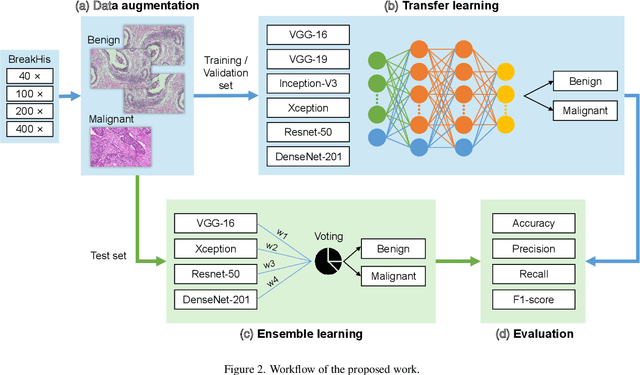
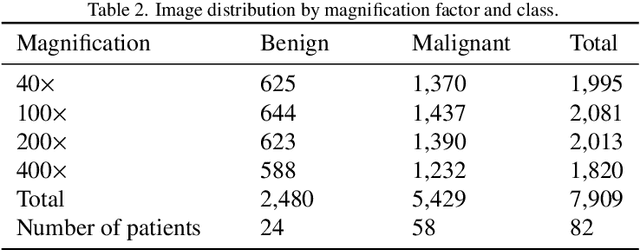
Abstract:Background: Breast cancer has the highest prevalence in women globally. The classification and diagnosis of breast cancer and its histopathological images have always been a hot spot of clinical concern. In Computer-Aided Diagnosis (CAD), traditional classification models mostly use a single network to extract features, which has significant limitations. On the other hand, many networks are trained and optimized on patient-level datasets, ignoring the application of lower-level data labels. Method: This paper proposes a deep ensemble model based on image-level labels for the binary classification of benign and malignant lesions of breast histopathological images. First, the BreakHis dataset is randomly divided into a training, validation and test set. Then, data augmentation techniques are used to balance the number of benign and malignant samples. Thirdly, considering the performance of transfer learning and the complementarity between each network, VGG-16, Xception, Resnet-50, DenseNet-201 are selected as the base classifiers. Result: In the ensemble network model with accuracy as the weight, the image-level binary classification achieves an accuracy of $98.90\%$. In order to verify the capabilities of our method, the latest Transformer and Multilayer Perception (MLP) models have been experimentally compared on the same dataset. Our model wins with a $5\%-20\%$ advantage, emphasizing the ensemble model's far-reaching significance in classification tasks. Conclusion: This research focuses on improving the model's classification performance with an ensemble algorithm. Transfer learning plays an essential role in small datasets, improving training speed and accuracy. Our model has outperformed many existing approaches in accuracy, providing a method for the field of auxiliary medical diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge