Haoyuan Chen
Improving the Predictability of the Madden-Julian Oscillation at Subseasonal Scales with Gaussian Process Models
May 21, 2025Abstract:The Madden--Julian Oscillation (MJO) is an influential climate phenomenon that plays a vital role in modulating global weather patterns. In spite of the improvement in MJO predictions made by machine learning algorithms, such as neural networks, most of them cannot provide the uncertainty levels in the MJO forecasts directly. To address this problem, we develop a nonparametric strategy based on Gaussian process (GP) models. We calibrate GPs using empirical correlations and we propose a posteriori covariance correction. Numerical experiments demonstrate that our model has better prediction skills than the ANN models for the first five lead days. Additionally, our posteriori covariance correction extends the probabilistic coverage by more than three weeks.
Gaussian Processes Sampling with Sparse Grids under Additive Schwarz Preconditioner
Aug 01, 2024Abstract:Gaussian processes (GPs) are widely used in non-parametric Bayesian modeling, and play an important role in various statistical and machine learning applications. In a variety tasks of uncertainty quantification, generating random sample paths of GPs is of interest. As GP sampling requires generating high-dimensional Gaussian random vectors, it is computationally challenging if a direct method, such as the Cholesky decomposition, is used. In this paper, we propose a scalable algorithm for sampling random realizations of the prior and posterior of GP models. The proposed algorithm leverages inducing points approximation with sparse grids, as well as additive Schwarz preconditioners, which reduce computational complexity, and ensure fast convergence. We demonstrate the efficacy and accuracy of the proposed method through a series of experiments and comparisons with other recent works.
Aggregation Models with Optimal Weights for Distributed Gaussian Processes
Aug 01, 2024Abstract:Gaussian process (GP) models have received increasingly attentions in recent years due to their superb prediction accuracy and modeling flexibility. To address the computational burdens of GP models for large-scale datasets, distributed learning for GPs are often adopted. Current aggregation models for distributed GPs are not time-efficient when incorporating correlations between GP experts. In this work, we propose a novel approach for aggregated prediction in distributed GPs. The technique is suitable for both the exact and sparse variational GPs. The proposed method incorporates correlations among experts, leading to better prediction accuracy with manageable computational requirements. As demonstrated by empirical studies, the proposed approach results in more stable predictions in less time than state-of-the-art consistent aggregation models.
MS-UNet-v2: Adaptive Denoising Method and Training Strategy for Medical Image Segmentation with Small Training Data
Sep 07, 2023



Abstract:Models based on U-like structures have improved the performance of medical image segmentation. However, the single-layer decoder structure of U-Net is too "thin" to exploit enough information, resulting in large semantic differences between the encoder and decoder parts. Things get worse if the number of training sets of data is not sufficiently large, which is common in medical image processing tasks where annotated data are more difficult to obtain than other tasks. Based on this observation, we propose a novel U-Net model named MS-UNet for the medical image segmentation task in this study. Instead of the single-layer U-Net decoder structure used in Swin-UNet and TransUnet, we specifically design a multi-scale nested decoder based on the Swin Transformer for U-Net. The proposed multi-scale nested decoder structure allows the feature mapping between the decoder and encoder to be semantically closer, thus enabling the network to learn more detailed features. In addition, we propose a novel edge loss and a plug-and-play fine-tuning Denoising module, which not only effectively improves the segmentation performance of MS-UNet, but could also be applied to other models individually. Experimental results show that MS-UNet could effectively improve the network performance with more efficient feature learning capability and exhibit more advanced performance, especially in the extreme case with a small amount of training data, and the proposed Edge loss and Denoising module could significantly enhance the segmentation performance of MS-UNet.
Representing Additive Gaussian Processes by Sparse Matrices
Apr 29, 2023Abstract:Among generalized additive models, additive Mat\'ern Gaussian Processes (GPs) are one of the most popular for scalable high-dimensional problems. Thanks to their additive structure and stochastic differential equation representation, back-fitting-based algorithms can reduce the time complexity of computing the posterior mean from $O(n^3)$ to $O(n\log n)$ time where $n$ is the data size. However, generalizing these algorithms to efficiently compute the posterior variance and maximum log-likelihood remains an open problem. In this study, we demonstrate that for Additive Mat\'ern GPs, not only the posterior mean, but also the posterior variance, log-likelihood, and gradient of these three functions can be represented by formulas involving only sparse matrices and sparse vectors. We show how to use these sparse formulas to generalize back-fitting-based algorithms to efficiently compute the posterior mean, posterior variance, log-likelihood, and gradient of these three functions for additive GPs, all in $O(n \log n)$ time. We apply our algorithms to Bayesian optimization and propose efficient algorithms for posterior updates, hyperparameters learning, and computations of the acquisition function and its gradient in Bayesian optimization. Given the posterior, our algorithms significantly reduce the time complexity of computing the acquisition function and its gradient from $O(n^2)$ to $O(\log n)$ for general learning rate, and even to $O(1)$ for small learning rate.
EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks
Dec 06, 2022



Abstract:Background and Purpose: Colorectal cancer is a common fatal malignancy, the fourth most common cancer in men, and the third most common cancer in women worldwide. Timely detection of cancer in its early stages is essential for treating the disease. Currently, there is a lack of datasets for histopathological image segmentation of rectal cancer, which often hampers the assessment accuracy when computer technology is used to aid in diagnosis. Methods: This present study provided a new publicly available Enteroscope Biopsy Histopathological Hematoxylin and Eosin Image Dataset for Image Segmentation Tasks (EBHI-Seg). To demonstrate the validity and extensiveness of EBHI-Seg, the experimental results for EBHI-Seg are evaluated using classical machine learning methods and deep learning methods. Results: The experimental results showed that deep learning methods had a better image segmentation performance when utilizing EBHI-Seg. The maximum accuracy of the Dice evaluation metric for the classical machine learning method is 0.948, while the Dice evaluation metric for the deep learning method is 0.965. Conclusion: This publicly available dataset contained 5,170 images of six types of tumor differentiation stages and the corresponding ground truth images. The dataset can provide researchers with new segmentation algorithms for medical diagnosis of colorectal cancer, which can be used in the clinical setting to help doctors and patients.
IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach
Jun 07, 2022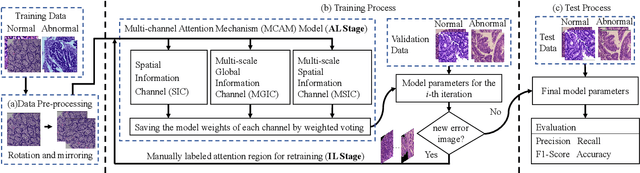
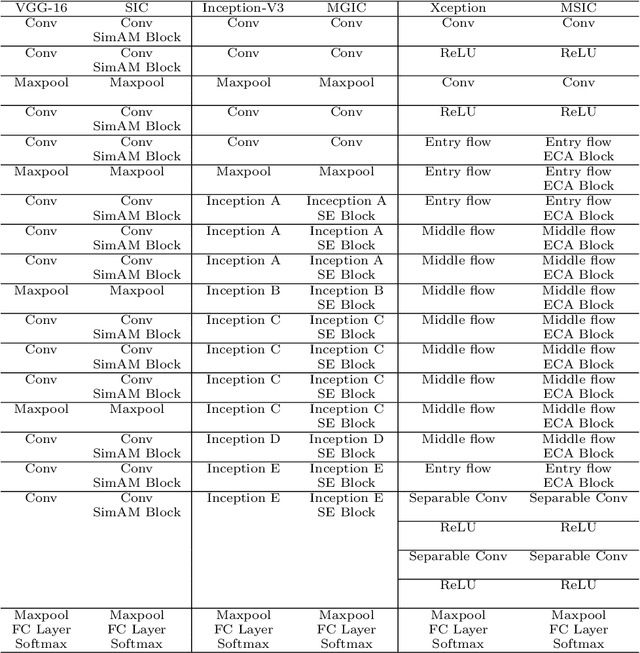
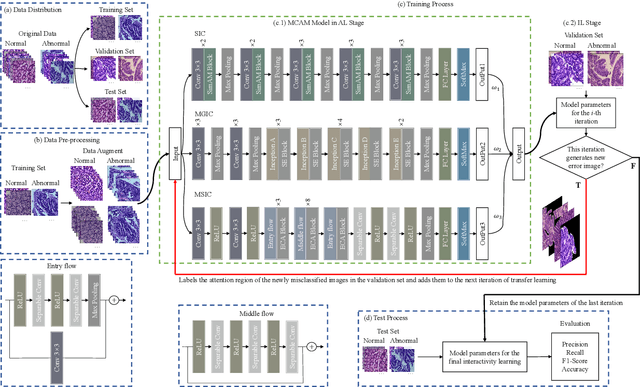
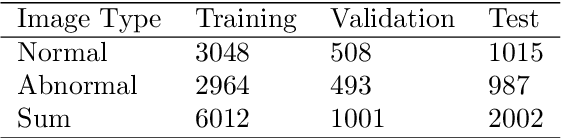
Abstract:In recent years, colorectal cancer has become one of the most significant diseases that endanger human health. Deep learning methods are increasingly important for the classification of colorectal histopathology images. However, existing approaches focus more on end-to-end automatic classification using computers rather than human-computer interaction. In this paper, we propose an IL-MCAM framework. It is based on attention mechanisms and interactive learning. The proposed IL-MCAM framework includes two stages: automatic learning (AL) and interactivity learning (IL). In the AL stage, a multi-channel attention mechanism model containing three different attention mechanism channels and convolutional neural networks is used to extract multi-channel features for classification. In the IL stage, the proposed IL-MCAM framework continuously adds misclassified images to the training set in an interactive approach, which improves the classification ability of the MCAM model. We carried out a comparison experiment on our dataset and an extended experiment on the HE-NCT-CRC-100K dataset to verify the performance of the proposed IL-MCAM framework, achieving classification accuracies of 98.98% and 99.77%, respectively. In addition, we conducted an ablation experiment and an interchangeability experiment to verify the ability and interchangeability of the three channels. The experimental results show that the proposed IL-MCAM framework has excellent performance in the colorectal histopathological image classification tasks.
CVM-Cervix: A Hybrid Cervical Pap-Smear Image Classification Framework Using CNN, Visual Transformer and Multilayer Perceptron
Jun 02, 2022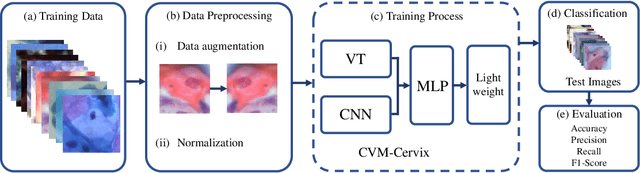

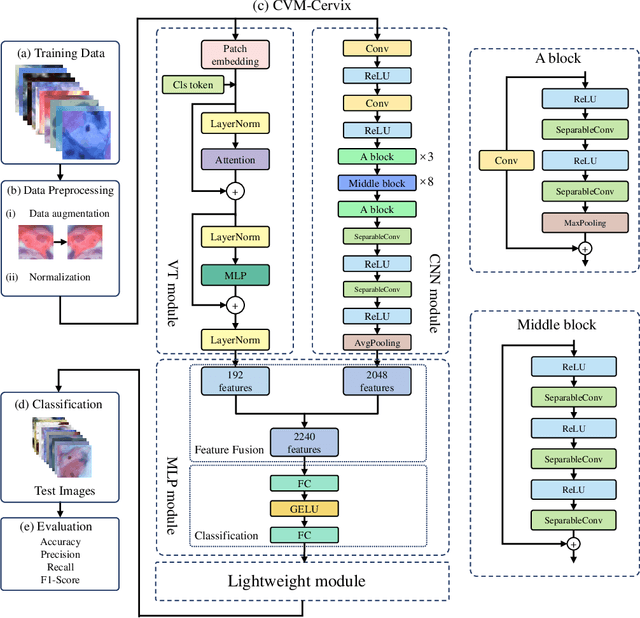

Abstract:Cervical cancer is the seventh most common cancer among all the cancers worldwide and the fourth most common cancer among women. Cervical cytopathology image classification is an important method to diagnose cervical cancer. Manual screening of cytopathology images is time-consuming and error-prone. The emergence of the automatic computer-aided diagnosis system solves this problem. This paper proposes a framework called CVM-Cervix based on deep learning to perform cervical cell classification tasks. It can analyze pap slides quickly and accurately. CVM-Cervix first proposes a Convolutional Neural Network module and a Visual Transformer module for local and global feature extraction respectively, then a Multilayer Perceptron module is designed to fuse the local and global features for the final classification. Experimental results show the effectiveness and potential of the proposed CVM-Cervix in the field of cervical Pap smear image classification. In addition, according to the practical needs of clinical work, we perform a lightweight post-processing to compress the model.
A Comparative Study of Gastric Histopathology Sub-size Image Classification: from Linear Regression to Visual Transformer
May 25, 2022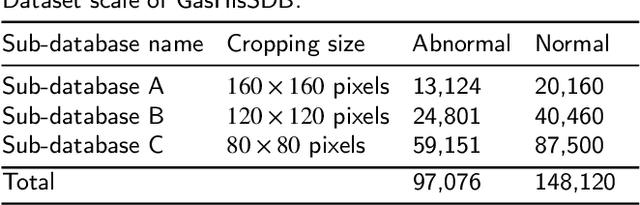
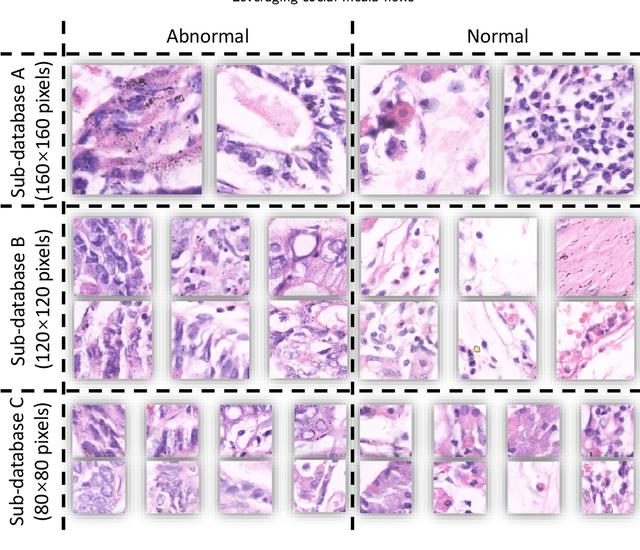
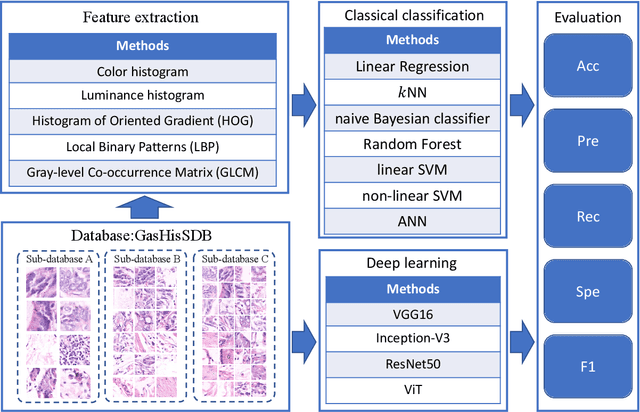
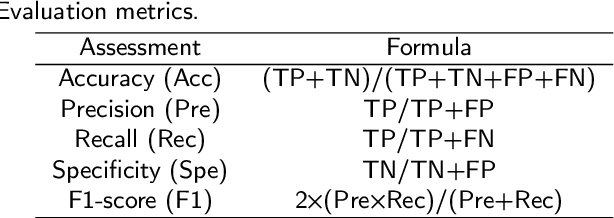
Abstract:Gastric cancer is the fifth most common cancer in the world. At the same time, it is also the fourth most deadly cancer. Early detection of cancer exists as a guide for the treatment of gastric cancer. Nowadays, computer technology has advanced rapidly to assist physicians in the diagnosis of pathological pictures of gastric cancer. Ensemble learning is a way to improve the accuracy of algorithms, and finding multiple learning models with complementarity types is the basis of ensemble learning. The complementarity of sub-size pathology image classifiers when machine performance is insufficient is explored in this experimental platform. We choose seven classical machine learning classifiers and four deep learning classifiers for classification experiments on the GasHisSDB database. Among them, classical machine learning algorithms extract five different image virtual features to match multiple classifier algorithms. For deep learning, we choose three convolutional neural network classifiers. In addition, we also choose a novel Transformer-based classifier. The experimental platform, in which a large number of classical machine learning and deep learning methods are performed, demonstrates that there are differences in the performance of different classifiers on GasHisSDB. Classical machine learning models exist for classifiers that classify Abnormal categories very well, while classifiers that excel in classifying Normal categories also exist. Deep learning models also exist with multiple models that can be complementarity. Suitable classifiers are selected for ensemble learning, when machine performance is insufficient. This experimental platform demonstrates that multiple classifiers are indeed complementarity and can improve the efficiency of ensemble learning. This can better assist doctors in diagnosis, improve the detection of gastric cancer, and increase the cure rate.
Application of Graph Based Features in Computer Aided Diagnosis for Histopathological Image Classification of Gastric Cancer
May 17, 2022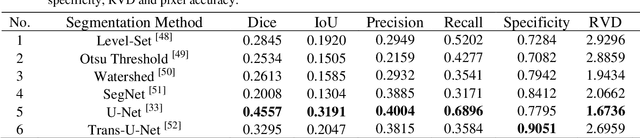
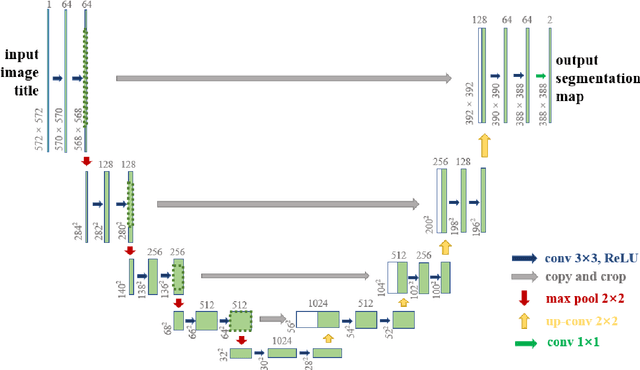

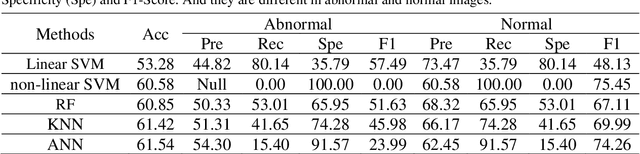
Abstract:The gold standard for gastric cancer detection is gastric histopathological image analysis, but there are certain drawbacks in the existing histopathological detection and diagnosis. In this paper, based on the study of computer aided diagnosis system, graph based features are applied to gastric cancer histopathology microscopic image analysis, and a classifier is used to classify gastric cancer cells from benign cells. Firstly, image segmentation is performed, and after finding the region, cell nuclei are extracted using the k-means method, the minimum spanning tree (MST) is drawn, and graph based features of the MST are extracted. The graph based features are then put into the classifier for classification. In this study, different segmentation methods are compared in the tissue segmentation stage, among which are Level-Set, Otsu thresholding, watershed, SegNet, U-Net and Trans-U-Net segmentation; Graph based features, Red, Green, Blue features, Grey-Level Co-occurrence Matrix features, Histograms of Oriented Gradient features and Local Binary Patterns features are compared in the feature extraction stage; Radial Basis Function (RBF) Support Vector Machine (SVM), Linear SVM, Artificial Neural Network, Random Forests, k-NearestNeighbor, VGG16, and Inception-V3 are compared in the classifier stage. It is found that using U-Net to segment tissue areas, then extracting graph based features, and finally using RBF SVM classifier gives the optimal results with 94.29%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge