Application of Graph Based Features in Computer Aided Diagnosis for Histopathological Image Classification of Gastric Cancer
Paper and Code
May 17, 2022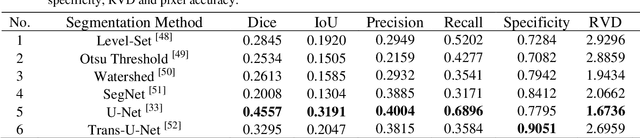
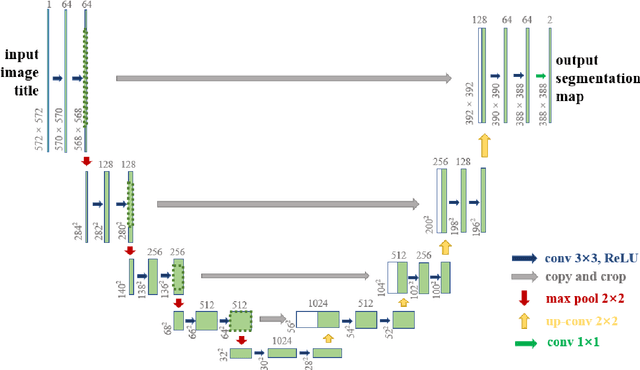

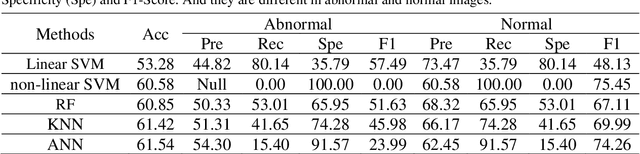
The gold standard for gastric cancer detection is gastric histopathological image analysis, but there are certain drawbacks in the existing histopathological detection and diagnosis. In this paper, based on the study of computer aided diagnosis system, graph based features are applied to gastric cancer histopathology microscopic image analysis, and a classifier is used to classify gastric cancer cells from benign cells. Firstly, image segmentation is performed, and after finding the region, cell nuclei are extracted using the k-means method, the minimum spanning tree (MST) is drawn, and graph based features of the MST are extracted. The graph based features are then put into the classifier for classification. In this study, different segmentation methods are compared in the tissue segmentation stage, among which are Level-Set, Otsu thresholding, watershed, SegNet, U-Net and Trans-U-Net segmentation; Graph based features, Red, Green, Blue features, Grey-Level Co-occurrence Matrix features, Histograms of Oriented Gradient features and Local Binary Patterns features are compared in the feature extraction stage; Radial Basis Function (RBF) Support Vector Machine (SVM), Linear SVM, Artificial Neural Network, Random Forests, k-NearestNeighbor, VGG16, and Inception-V3 are compared in the classifier stage. It is found that using U-Net to segment tissue areas, then extracting graph based features, and finally using RBF SVM classifier gives the optimal results with 94.29%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge