Shouliang Qi
An attention-based deep learning network for predicting Platinum resistance in ovarian cancer
Nov 08, 2023



Abstract:Background: Ovarian cancer is among the three most frequent gynecologic cancers globally. High-grade serous ovarian cancer (HGSOC) is the most common and aggressive histological type. Guided treatment for HGSOC typically involves platinum-based combination chemotherapy, necessitating an assessment of whether the patient is platinum-resistant. The purpose of this study is to propose a deep learning-based method to determine whether a patient is platinum-resistant using multimodal positron emission tomography/computed tomography (PET/CT) images. Methods: 289 patients with HGSOC were included in this study. An end-to-end SE-SPP-DenseNet model was built by adding Squeeze-Excitation Block (SE Block) and Spatial Pyramid Pooling Layer (SPPLayer) to Dense Convolutional Network (DenseNet). Multimodal data from PET/CT images of the regions of interest (ROI) were used to predict platinum resistance in patients. Results: Through five-fold cross-validation, SE-SPP-DenseNet achieved a high accuracy rate and an area under the curve (AUC) in predicting platinum resistance in patients, which were 92.6% and 0.93, respectively. The importance of incorporating SE Block and SPPLayer into the deep learning model, and considering multimodal data was substantiated by carrying out ablation studies and experiments with single modality data. Conclusions: The obtained classification results indicate that our proposed deep learning framework performs better in predicting platinum resistance in patients, which can help gynecologists make better treatment decisions. Keywords: PET/CT, CNN, SE Block, SPP Layer, Platinum resistance, Ovarian cancer
NL-CS Net: Deep Learning with Non-Local Prior for Image Compressive Sensing
May 06, 2023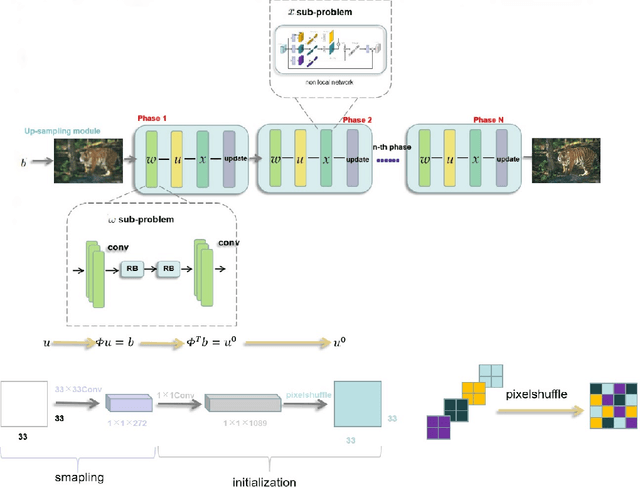
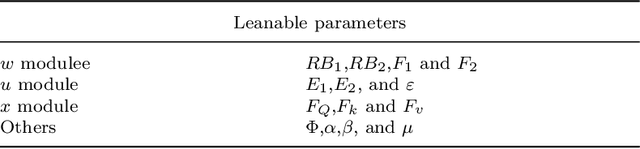
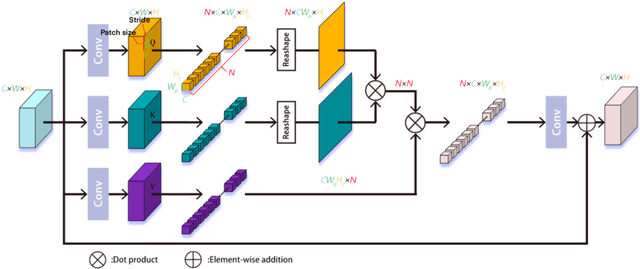
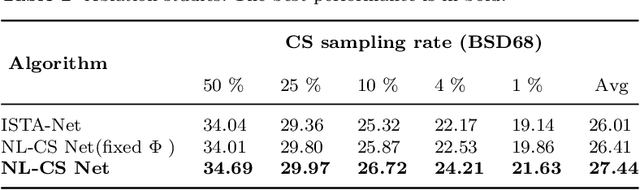
Abstract:Deep learning has been applied to compressive sensing (CS) of images successfully in recent years. However, existing network-based methods are often trained as the black box, in which the lack of prior knowledge is often the bottleneck for further performance improvement. To overcome this drawback, this paper proposes a novel CS method using non-local prior which combines the interpretability of the traditional optimization methods with the speed of network-based methods, called NL-CS Net. We unroll each phase from iteration of the augmented Lagrangian method solving non-local and sparse regularized optimization problem by a network. NL-CS Net is composed of the up-sampling module and the recovery module. In the up-sampling module, we use learnable up-sampling matrix instead of a predefined one. In the recovery module, patch-wise non-local network is employed to capture long-range feature correspondences. Important parameters involved (e.g. sampling matrix, nonlinear transforms, shrinkage thresholds, step size, $etc.$) are learned end-to-end, rather than hand-crafted. Furthermore, to facilitate practical implementation, orthogonal and binary constraints on the sampling matrix are simultaneously adopted. Extensive experiments on natural images and magnetic resonance imaging (MRI) demonstrate that the proposed method outperforms the state-of-the-art methods while maintaining great interpretability and speed.
Two-stage Contextual Transformer-based Convolutional Neural Network for Airway Extraction from CT Images
Dec 15, 2022Abstract:Accurate airway extraction from computed tomography (CT) images is a critical step for planning navigation bronchoscopy and quantitative assessment of airway-related chronic obstructive pulmonary disease (COPD). The existing methods are challenging to sufficiently segment the airway, especially the high-generation airway, with the constraint of the limited label and cannot meet the clinical use in COPD. We propose a novel two-stage 3D contextual transformer-based U-Net for airway segmentation using CT images. The method consists of two stages, performing initial and refined airway segmentation. The two-stage model shares the same subnetwork with different airway masks as input. Contextual transformer block is performed both in the encoder and decoder path of the subnetwork to finish high-quality airway segmentation effectively. In the first stage, the total airway mask and CT images are provided to the subnetwork, and the intrapulmonary airway mask and corresponding CT scans to the subnetwork in the second stage. Then the predictions of the two-stage method are merged as the final prediction. Extensive experiments were performed on in-house and multiple public datasets. Quantitative and qualitative analysis demonstrate that our proposed method extracted much more branches and lengths of the tree while accomplishing state-of-the-art airway segmentation performance. The code is available at https://github.com/zhaozsq/airway_segmentation.
EBHI-Seg: A Novel Enteroscope Biopsy Histopathological Haematoxylin and Eosin Image Dataset for Image Segmentation Tasks
Dec 06, 2022



Abstract:Background and Purpose: Colorectal cancer is a common fatal malignancy, the fourth most common cancer in men, and the third most common cancer in women worldwide. Timely detection of cancer in its early stages is essential for treating the disease. Currently, there is a lack of datasets for histopathological image segmentation of rectal cancer, which often hampers the assessment accuracy when computer technology is used to aid in diagnosis. Methods: This present study provided a new publicly available Enteroscope Biopsy Histopathological Hematoxylin and Eosin Image Dataset for Image Segmentation Tasks (EBHI-Seg). To demonstrate the validity and extensiveness of EBHI-Seg, the experimental results for EBHI-Seg are evaluated using classical machine learning methods and deep learning methods. Results: The experimental results showed that deep learning methods had a better image segmentation performance when utilizing EBHI-Seg. The maximum accuracy of the Dice evaluation metric for the classical machine learning method is 0.948, while the Dice evaluation metric for the deep learning method is 0.965. Conclusion: This publicly available dataset contained 5,170 images of six types of tumor differentiation stages and the corresponding ground truth images. The dataset can provide researchers with new segmentation algorithms for medical diagnosis of colorectal cancer, which can be used in the clinical setting to help doctors and patients.
Subspace Nonnegative Matrix Factorization for Feature Representation
Apr 18, 2022
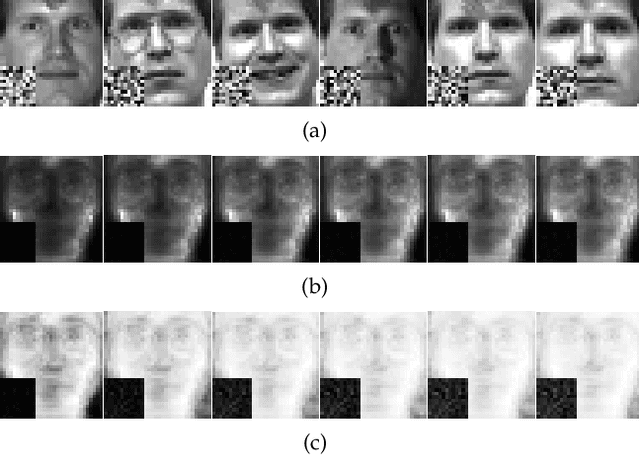
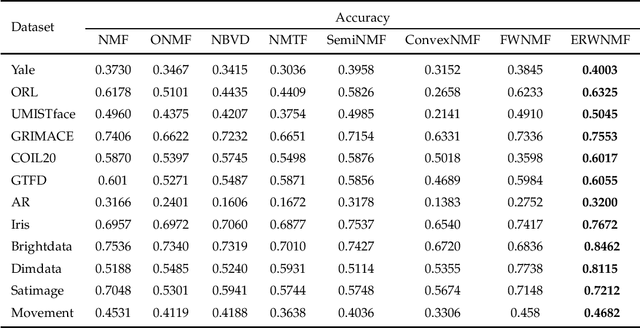
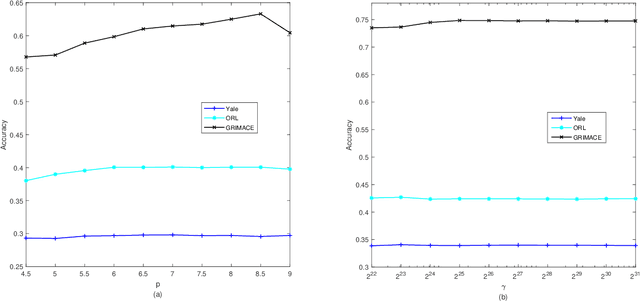
Abstract:Traditional nonnegative matrix factorization (NMF) learns a new feature representation on the whole data space, which means treating all features equally. However, a subspace is often sufficient for accurate representation in practical applications, and redundant features can be invalid or even harmful. For example, if a camera has some sensors destroyed, then the corresponding pixels in the photos from this camera are not helpful to identify the content, which means only the subspace consisting of remaining pixels is worthy of attention. This paper proposes a new NMF method by introducing adaptive weights to identify key features in the original space so that only a subspace involves generating the new representation. Two strategies are proposed to achieve this: the fuzzier weighted technique and entropy regularized weighted technique, both of which result in an iterative solution with a simple form. Experimental results on several real-world datasets demonstrated that the proposed methods can generate a more accurate feature representation than existing methods. The code developed in this study is available at https://github.com/WNMF1/FWNMF-ERWNMF.
Improving the Level of Autism Discrimination through GraphRNN Link Prediction
Feb 19, 2022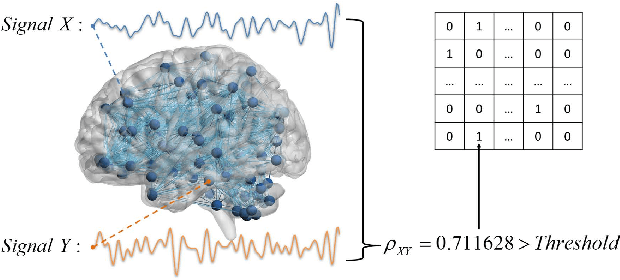
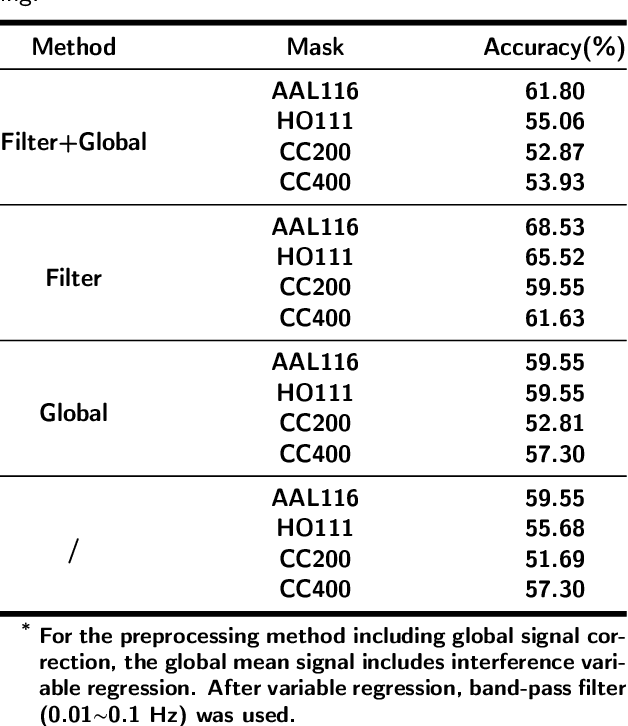
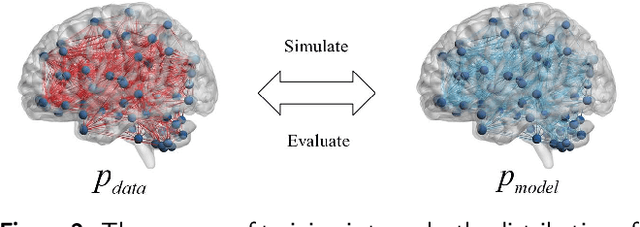
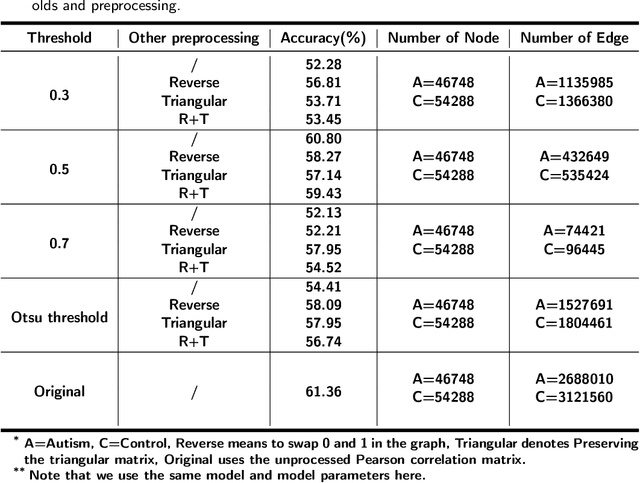
Abstract:Dataset is the key of deep learning in Autism disease research. However, due to the few quantity and heterogeneity of samples in current dataset, for example ABIDE (Autism Brain Imaging Data Exchange), the recognition research is not effective enough. Previous studies mostly focused on optimizing feature selection methods and data reinforcement to improve accuracy. This paper is based on the latter technique, which learns the edge distribution of real brain network through GraphRNN, and generates the synthetic data which has incentive effect on the discriminant model. The experimental results show that the combination of original and synthetic data greatly improves the discrimination of the neural network. For instance, the most significant effect is the 50-layer ResNet, and the best generation model is GraphRNN, which improves the accuracy by 32.51% compared with the model reference experiment without generation data reinforcement. Because the generated data comes from the learned edge connection distribution of Autism patients and typical controls functional connectivity, but it has better effect than the original data, which has constructive significance for further understanding of disease mechanism and development.
An Entropy Weighted Nonnegative Matrix Factorization Algorithm for Feature Representation
Nov 27, 2021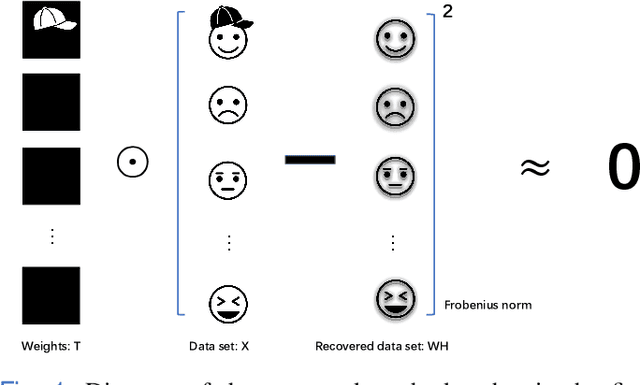
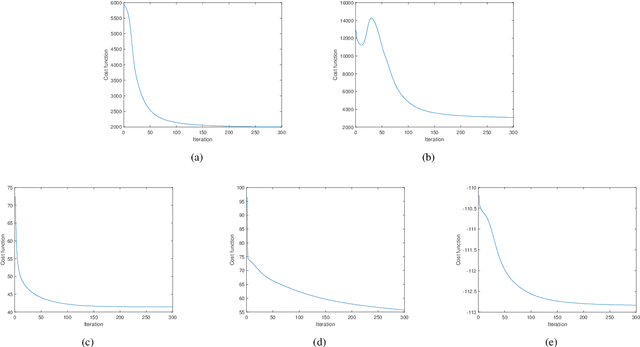

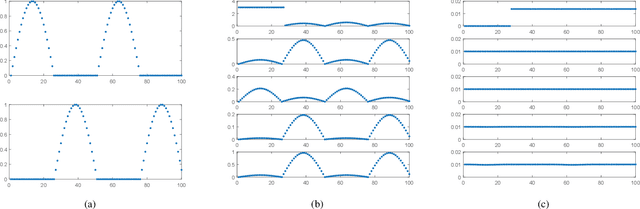
Abstract:Nonnegative matrix factorization (NMF) has been widely used to learn low-dimensional representations of data. However, NMF pays the same attention to all attributes of a data point, which inevitably leads to inaccurate representation. For example, in a human-face data set, if an image contains a hat on the head, the hat should be removed or the importance of its corresponding attributes should be decreased during matrix factorizing. This paper proposes a new type of NMF called entropy weighted NMF (EWNMF), which uses an optimizable weight for each attribute of each data point to emphasize their importance. This process is achieved by adding an entropy regularizer to the cost function and then using the Lagrange multiplier method to solve the problem. Experimental results with several data sets demonstrate the feasibility and effectiveness of the proposed method. We make our code available at https://github.com/Poisson-EM/Entropy-weighted-NMF.
EMDS-7: Environmental Microorganism Image Dataset Seventh Version for Multiple Object Detection Evaluation
Oct 28, 2021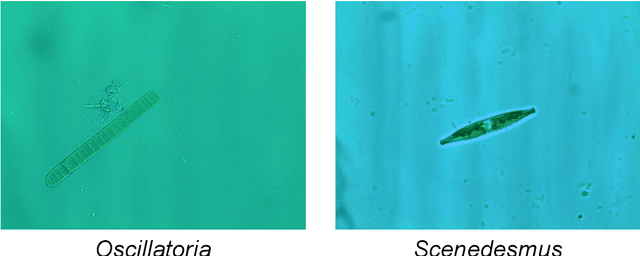
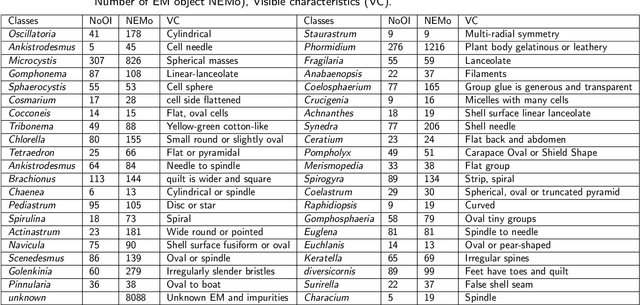

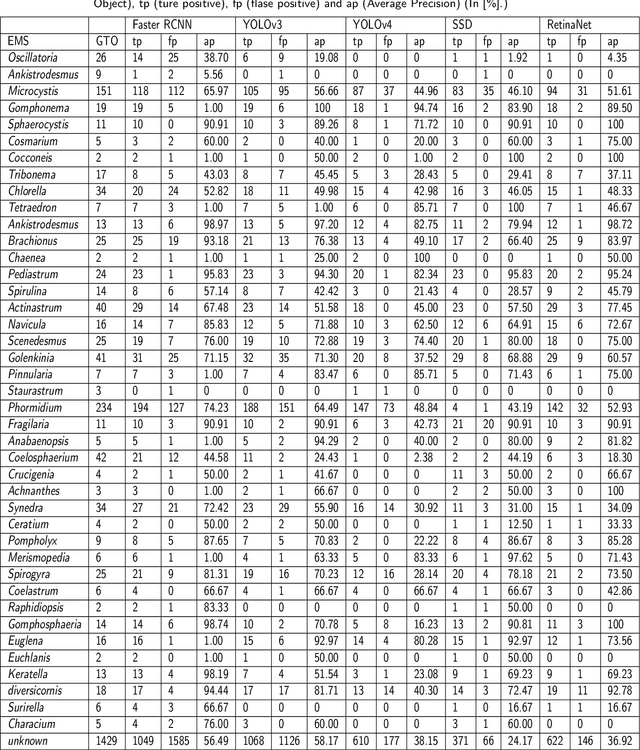
Abstract:The Environmental Microorganism Image Dataset Seventh Version (EMDS-7) is a microscopic image data set, including the original Environmental Microorganism images (EMs) and the corresponding object labeling files in ".XML" format file. The EMDS-7 data set consists of 41 types of EMs, which has a total of 2365 images and 13216 labeled objects. The EMDS-7 database mainly focuses on the object detection. In order to prove the effectiveness of EMDS-7, we select the most commonly used deep learning methods (Faster-RCNN, YOLOv3, YOLOv4, SSD and RetinaNet) and evaluation indices for testing and evaluation. EMDS-7 is freely published for non-commercial purpose at: https://figshare.com/articles/dataset/EMDS-7_DataSet/16869571
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge