Yuanjing Feng
Zhejiang University of Technology, Hangzhou, China
Convergent and divergent connectivity patterns of the arcuate fasciculus in macaques and humans
Jun 24, 2025Abstract:The organization and connectivity of the arcuate fasciculus (AF) in nonhuman primates remain contentious, especially concerning how its anatomy diverges from that of humans. Here, we combined cross-scale single-neuron tracing - using viral-based genetic labeling and fluorescence micro-optical sectioning tomography in macaques (n = 4; age 3 - 11 years) - with whole-brain tractography from 11.7T diffusion MRI. Complemented by spectral embedding analysis of 7.0T MRI in humans, we performed a comparative connectomic analysis of the AF across species. We demonstrate that the macaque AF originates in the temporal-parietal cortex, traverses the auditory cortex and parietal operculum, and projects into prefrontal regions. In contrast, the human AF exhibits greater expansion into the middle temporal gyrus and stronger prefrontal and parietal operculum connectivity - divergences quantified by Kullback-Leibler analysis that likely underpin the evolutionary specialization of human language networks. These interspecies differences - particularly the human AF's broader temporal integration and strengthened frontoparietal linkages - suggest a connectivity-based substrate for the emergence of advanced language processing unique to humans. Furthermore, our findings offer a neuroanatomical framework for understanding AF-related disorders such as aphasia and dyslexia, where aberrant connectivity disrupts language function.
Cross-Sequence Semi-Supervised Learning for Multi-Parametric MRI-Based Visual Pathway Delineation
May 26, 2025Abstract:Accurately delineating the visual pathway (VP) is crucial for understanding the human visual system and diagnosing related disorders. Exploring multi-parametric MR imaging data has been identified as an important way to delineate VP. However, due to the complex cross-sequence relationships, existing methods cannot effectively model the complementary information from different MRI sequences. In addition, these existing methods heavily rely on large training data with labels, which is labor-intensive and time-consuming to obtain. In this work, we propose a novel semi-supervised multi-parametric feature decomposition framework for VP delineation. Specifically, a correlation-constrained feature decomposition (CFD) is designed to handle the complex cross-sequence relationships by capturing the unique characteristics of each MRI sequence and easing the multi-parametric information fusion process. Furthermore, a consistency-based sample enhancement (CSE) module is developed to address the limited labeled data issue, by generating and promoting meaningful edge information from unlabeled data. We validate our framework using two public datasets, and one in-house Multi-Shell Diffusion MRI (MDM) dataset. Experimental results demonstrate the superiority of our approach in terms of delineation performance when compared to seven state-of-the-art approaches.
An Arbitrary-Modal Fusion Network for Volumetric Cranial Nerves Tract Segmentation
May 05, 2025Abstract:The segmentation of cranial nerves (CNs) tract provides a valuable quantitative tool for the analysis of the morphology and trajectory of individual CNs. Multimodal CNs tract segmentation networks, e.g., CNTSeg, which combine structural Magnetic Resonance Imaging (MRI) and diffusion MRI, have achieved promising segmentation performance. However, it is laborious or even infeasible to collect complete multimodal data in clinical practice due to limitations in equipment, user privacy, and working conditions. In this work, we propose a novel arbitrary-modal fusion network for volumetric CNs tract segmentation, called CNTSeg-v2, which trains one model to handle different combinations of available modalities. Instead of directly combining all the modalities, we select T1-weighted (T1w) images as the primary modality due to its simplicity in data acquisition and contribution most to the results, which supervises the information selection of other auxiliary modalities. Our model encompasses an Arbitrary-Modal Collaboration Module (ACM) designed to effectively extract informative features from other auxiliary modalities, guided by the supervision of T1w images. Meanwhile, we construct a Deep Distance-guided Multi-stage (DDM) decoder to correct small errors and discontinuities through signed distance maps to improve segmentation accuracy. We evaluate our CNTSeg-v2 on the Human Connectome Project (HCP) dataset and the clinical Multi-shell Diffusion MRI (MDM) dataset. Extensive experimental results show that our CNTSeg-v2 achieves state-of-the-art segmentation performance, outperforming all competing methods.
DeepMpMRI: Tensor-decomposition Regularized Learning for Fast and High-Fidelity Multi-Parametric Microstructural MR Imaging
May 06, 2024Abstract:Deep learning has emerged as a promising approach for learning the nonlinear mapping between diffusion-weighted MR images and tissue parameters, which enables automatic and deep understanding of the brain microstructures. However, the efficiency and accuracy in the multi-parametric estimations are still limited since previous studies tend to estimate multi-parametric maps with dense sampling and isolated signal modeling. This paper proposes DeepMpMRI, a unified framework for fast and high-fidelity multi-parametric estimation from various diffusion models using sparsely sampled q-space data. DeepMpMRI is equipped with a newly designed tensor-decomposition-based regularizer to effectively capture fine details by exploiting the correlation across parameters. In addition, we introduce a Nesterov-based adaptive learning algorithm that optimizes the regularization parameter dynamically to enhance the performance. DeepMpMRI is an extendable framework capable of incorporating flexible network architecture. Experimental results demonstrate the superiority of our approach over 5 state-of-the-art methods in simultaneously estimating multi-parametric maps for various diffusion models with fine-grained details both quantitatively and qualitatively, achieving 4.5 - 22.5$\times$ acceleration compared to the dense sampling of a total of 270 diffusion gradients.
Anatomy-guided fiber trajectory distribution estimation for cranial nerves tractography
Feb 29, 2024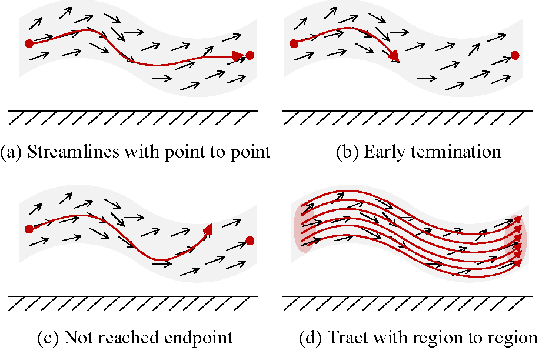
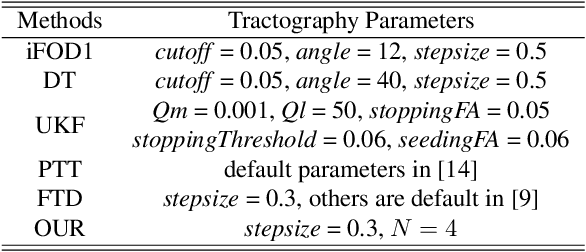
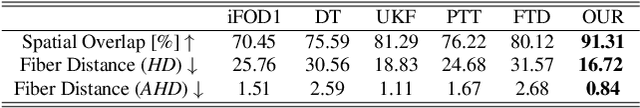
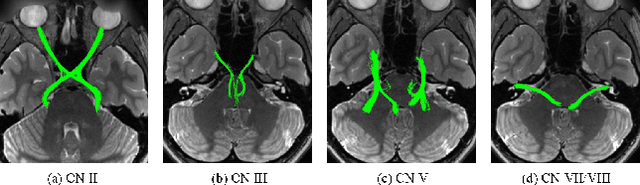
Abstract:Diffusion MRI tractography is an important tool for identifying and analyzing the intracranial course of cranial nerves (CNs). However, the complex environment of the skull base leads to ambiguous spatial correspondence between diffusion directions and fiber geometry, and existing diffusion tractography methods of CNs identification are prone to producing erroneous trajectories and missing true positive connections. To overcome the above challenge, we propose a novel CNs identification framework with anatomy-guided fiber trajectory distribution, which incorporates anatomical shape prior knowledge during the process of CNs tracing to build diffusion tensor vector fields. We introduce higher-order streamline differential equations for continuous flow field representations to directly characterize the fiber trajectory distribution of CNs from the tract-based level. The experimental results on the vivo HCP dataset and the clinical MDM dataset demonstrate that the proposed method reduces false-positive fiber production compared to competing methods and produces reconstructed CNs (i.e. CN II, CN III, CN V, and CN VII/VIII) that are judged to better correspond to the known anatomy.
LESEN: Label-Efficient deep learning for Multi-parametric MRI-based Visual Pathway Segmentation
Jan 03, 2024Abstract:Recent research has shown the potential of deep learning in multi-parametric MRI-based visual pathway (VP) segmentation. However, obtaining labeled data for training is laborious and time-consuming. Therefore, it is crucial to develop effective algorithms in situations with limited labeled samples. In this work, we propose a label-efficient deep learning method with self-ensembling (LESEN). LESEN incorporates supervised and unsupervised losses, enabling the student and teacher models to mutually learn from each other, forming a self-ensembling mean teacher framework. Additionally, we introduce a reliable unlabeled sample selection (RUSS) mechanism to further enhance LESEN's effectiveness. Our experiments on the human connectome project (HCP) dataset demonstrate the superior performance of our method when compared to state-of-the-art techniques, advancing multimodal VP segmentation for comprehensive analysis in clinical and research settings. The implementation code will be available at: https://github.com/aldiak/Semi-Supervised-Multimodal-Visual-Pathway- Delineation.
Modality Exchange Network for Retinogeniculate Visual Pathway Segmentation
Jan 03, 2024Abstract:Accurate segmentation of the retinogeniculate visual pathway (RGVP) aids in the diagnosis and treatment of visual disorders by identifying disruptions or abnormalities within the pathway. However, the complex anatomical structure and connectivity of RGVP make it challenging to achieve accurate segmentation. In this study, we propose a novel Modality Exchange Network (ME-Net) that effectively utilizes multi-modal magnetic resonance (MR) imaging information to enhance RGVP segmentation. Our ME-Net has two main contributions. Firstly, we introduce an effective multi-modal soft-exchange technique. Specifically, we design a channel and spatially mixed attention module to exchange modality information between T1-weighted and fractional anisotropy MR images. Secondly, we propose a cross-fusion module that further enhances the fusion of information between the two modalities. Experimental results demonstrate that our method outperforms existing state-of-the-art approaches in terms of RGVP segmentation performance.
Bundle-specific Tractogram Distribution Estimation Using Higher-order Streamline Differential Equation
Jul 06, 2023Abstract:Tractography traces the peak directions extracted from fiber orientation distribution (FOD) suffering from ambiguous spatial correspondences between diffusion directions and fiber geometry, which is prone to producing erroneous tracks while missing true positive connections. The peaks-based tractography methods 'locally' reconstructed streamlines in 'single to single' manner, thus lacking of global information about the trend of the whole fiber bundle. In this work, we propose a novel tractography method based on a bundle-specific tractogram distribution function by using a higher-order streamline differential equation, which reconstructs the streamline bundles in 'cluster to cluster' manner. A unified framework for any higher-order streamline differential equation is presented to describe the fiber bundles with disjoint streamlines defined based on the diffusion tensor vector field. At the global level, the tractography process is simplified as the estimation of bundle-specific tractogram distribution (BTD) coefficients by minimizing the energy optimization model, and is used to characterize the relations between BTD and diffusion tensor vector under the prior guidance by introducing the tractogram bundle information to provide anatomic priors. Experiments are performed on simulated Hough, Sine, Circle data, ISMRM 2015 Tractography Challenge data, FiberCup data, and in vivo data from the Human Connectome Project (HCP) data for qualitative and quantitative evaluation. The results demonstrate that our approach can reconstruct the complex global fiber bundles directly. BTD reduces the error deviation and accumulation at the local level and shows better results in reconstructing long-range, twisting, and large fanning tracts.
Reconstructing the somatotopic organization of the corticospinal tract remains a challenge for modern tractography methods
Jun 15, 2023Abstract:The corticospinal tract (CST) is a critically important white matter fiber tract in the human brain that enables control of voluntary movements of the body. Diffusion MRI tractography is the only method that enables the study of the anatomy and variability of the CST pathway in human health. In this work, we explored the performance of six widely used tractography methods for reconstructing the CST and its somatotopic organization. We perform experiments using diffusion MRI data from the Human Connectome Project. Four quantitative measurements including reconstruction rate, the WM-GM interface coverage, anatomical distribution of streamlines, and correlation with cortical volumes to assess the advantages and limitations of each method. Overall, we conclude that while current tractography methods have made progress toward the well-known challenge of improving the reconstruction of the lateral projections of the CST, the overall problem of performing a comprehensive CST reconstruction, including clinically important projections in the lateral (hand and face area) and medial portions (leg area), remains an important challenge for diffusion MRI tractography.
DeepRGVP: A Novel Microstructure-Informed Supervised Contrastive Learning Framework for Automated Identification Of The Retinogeniculate Pathway Using dMRI Tractography
Nov 15, 2022



Abstract:The retinogeniculate pathway (RGVP) is responsible for carrying visual information from the retina to the lateral geniculate nucleus. Identification and visualization of the RGVP are important in studying the anatomy of the visual system and can inform treatment of related brain diseases. Diffusion MRI (dMRI) tractography is an advanced imaging method that uniquely enables in vivo mapping of the 3D trajectory of the RGVP. Currently, identification of the RGVP from tractography data relies on expert (manual) selection of tractography streamlines, which is time-consuming, has high clinical and expert labor costs, and affected by inter-observer variability. In this paper, we present what we believe is the first deep learning framework, namely DeepRGVP, to enable fast and accurate identification of the RGVP from dMRI tractography data. We design a novel microstructure-informed supervised contrastive learning method that leverages both streamline label and tissue microstructure information to determine positive and negative pairs. We propose a simple and successful streamline-level data augmentation method to address highly imbalanced training data, where the number of RGVP streamlines is much lower than that of non-RGVP streamlines. We perform comparisons with several state-of-the-art deep learning methods that were designed for tractography parcellation, and we show superior RGVP identification results using DeepRGVP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge