Qingrun Zeng
Convergent and divergent connectivity patterns of the arcuate fasciculus in macaques and humans
Jun 24, 2025Abstract:The organization and connectivity of the arcuate fasciculus (AF) in nonhuman primates remain contentious, especially concerning how its anatomy diverges from that of humans. Here, we combined cross-scale single-neuron tracing - using viral-based genetic labeling and fluorescence micro-optical sectioning tomography in macaques (n = 4; age 3 - 11 years) - with whole-brain tractography from 11.7T diffusion MRI. Complemented by spectral embedding analysis of 7.0T MRI in humans, we performed a comparative connectomic analysis of the AF across species. We demonstrate that the macaque AF originates in the temporal-parietal cortex, traverses the auditory cortex and parietal operculum, and projects into prefrontal regions. In contrast, the human AF exhibits greater expansion into the middle temporal gyrus and stronger prefrontal and parietal operculum connectivity - divergences quantified by Kullback-Leibler analysis that likely underpin the evolutionary specialization of human language networks. These interspecies differences - particularly the human AF's broader temporal integration and strengthened frontoparietal linkages - suggest a connectivity-based substrate for the emergence of advanced language processing unique to humans. Furthermore, our findings offer a neuroanatomical framework for understanding AF-related disorders such as aphasia and dyslexia, where aberrant connectivity disrupts language function.
An Arbitrary-Modal Fusion Network for Volumetric Cranial Nerves Tract Segmentation
May 05, 2025Abstract:The segmentation of cranial nerves (CNs) tract provides a valuable quantitative tool for the analysis of the morphology and trajectory of individual CNs. Multimodal CNs tract segmentation networks, e.g., CNTSeg, which combine structural Magnetic Resonance Imaging (MRI) and diffusion MRI, have achieved promising segmentation performance. However, it is laborious or even infeasible to collect complete multimodal data in clinical practice due to limitations in equipment, user privacy, and working conditions. In this work, we propose a novel arbitrary-modal fusion network for volumetric CNs tract segmentation, called CNTSeg-v2, which trains one model to handle different combinations of available modalities. Instead of directly combining all the modalities, we select T1-weighted (T1w) images as the primary modality due to its simplicity in data acquisition and contribution most to the results, which supervises the information selection of other auxiliary modalities. Our model encompasses an Arbitrary-Modal Collaboration Module (ACM) designed to effectively extract informative features from other auxiliary modalities, guided by the supervision of T1w images. Meanwhile, we construct a Deep Distance-guided Multi-stage (DDM) decoder to correct small errors and discontinuities through signed distance maps to improve segmentation accuracy. We evaluate our CNTSeg-v2 on the Human Connectome Project (HCP) dataset and the clinical Multi-shell Diffusion MRI (MDM) dataset. Extensive experimental results show that our CNTSeg-v2 achieves state-of-the-art segmentation performance, outperforming all competing methods.
MICCAI-CDMRI 2023 QuantConn Challenge Findings on Achieving Robust Quantitative Connectivity through Harmonized Preprocessing of Diffusion MRI
Nov 14, 2024



Abstract:White matter alterations are increasingly implicated in neurological diseases and their progression. International-scale studies use diffusion-weighted magnetic resonance imaging (DW-MRI) to qualitatively identify changes in white matter microstructure and connectivity. Yet, quantitative analysis of DW-MRI data is hindered by inconsistencies stemming from varying acquisition protocols. There is a pressing need to harmonize the preprocessing of DW-MRI datasets to ensure the derivation of robust quantitative diffusion metrics across acquisitions. In the MICCAI-CDMRI 2023 QuantConn challenge, participants were provided raw data from the same individuals collected on the same scanner but with two different acquisitions and tasked with preprocessing the DW-MRI to minimize acquisition differences while retaining biological variation. Submissions are evaluated on the reproducibility and comparability of cross-acquisition bundle-wise microstructure measures, bundle shape features, and connectomics. The key innovations of the QuantConn challenge are that (1) we assess bundles and tractography in the context of harmonization for the first time, (2) we assess connectomics in the context of harmonization for the first time, and (3) we have 10x additional subjects over prior harmonization challenge, MUSHAC and 100x over SuperMUDI. We find that bundle surface area, fractional anisotropy, connectome assortativity, betweenness centrality, edge count, modularity, nodal strength, and participation coefficient measures are most biased by acquisition and that machine learning voxel-wise correction, RISH mapping, and NeSH methods effectively reduce these biases. In addition, microstructure measures AD, MD, RD, bundle length, connectome density, efficiency, and path length are least biased by these acquisition differences.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024/019
Anatomy-guided fiber trajectory distribution estimation for cranial nerves tractography
Feb 29, 2024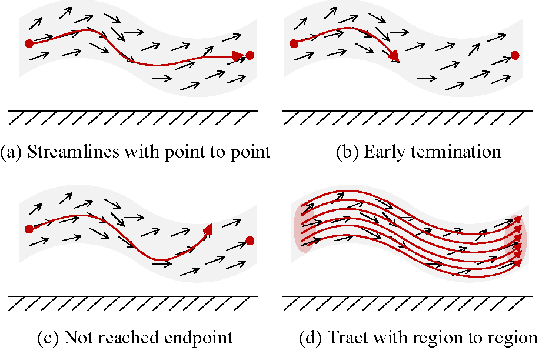
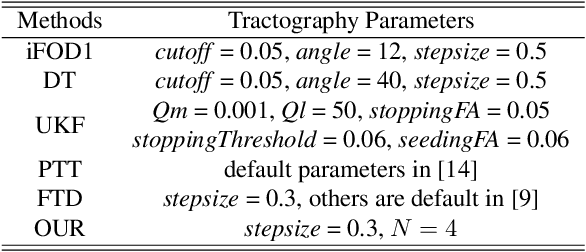
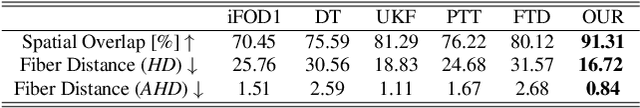
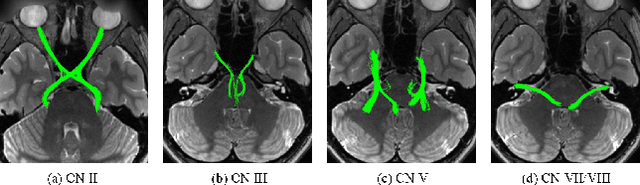
Abstract:Diffusion MRI tractography is an important tool for identifying and analyzing the intracranial course of cranial nerves (CNs). However, the complex environment of the skull base leads to ambiguous spatial correspondence between diffusion directions and fiber geometry, and existing diffusion tractography methods of CNs identification are prone to producing erroneous trajectories and missing true positive connections. To overcome the above challenge, we propose a novel CNs identification framework with anatomy-guided fiber trajectory distribution, which incorporates anatomical shape prior knowledge during the process of CNs tracing to build diffusion tensor vector fields. We introduce higher-order streamline differential equations for continuous flow field representations to directly characterize the fiber trajectory distribution of CNs from the tract-based level. The experimental results on the vivo HCP dataset and the clinical MDM dataset demonstrate that the proposed method reduces false-positive fiber production compared to competing methods and produces reconstructed CNs (i.e. CN II, CN III, CN V, and CN VII/VIII) that are judged to better correspond to the known anatomy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge