Guoqiang Xie
An Arbitrary-Modal Fusion Network for Volumetric Cranial Nerves Tract Segmentation
May 05, 2025Abstract:The segmentation of cranial nerves (CNs) tract provides a valuable quantitative tool for the analysis of the morphology and trajectory of individual CNs. Multimodal CNs tract segmentation networks, e.g., CNTSeg, which combine structural Magnetic Resonance Imaging (MRI) and diffusion MRI, have achieved promising segmentation performance. However, it is laborious or even infeasible to collect complete multimodal data in clinical practice due to limitations in equipment, user privacy, and working conditions. In this work, we propose a novel arbitrary-modal fusion network for volumetric CNs tract segmentation, called CNTSeg-v2, which trains one model to handle different combinations of available modalities. Instead of directly combining all the modalities, we select T1-weighted (T1w) images as the primary modality due to its simplicity in data acquisition and contribution most to the results, which supervises the information selection of other auxiliary modalities. Our model encompasses an Arbitrary-Modal Collaboration Module (ACM) designed to effectively extract informative features from other auxiliary modalities, guided by the supervision of T1w images. Meanwhile, we construct a Deep Distance-guided Multi-stage (DDM) decoder to correct small errors and discontinuities through signed distance maps to improve segmentation accuracy. We evaluate our CNTSeg-v2 on the Human Connectome Project (HCP) dataset and the clinical Multi-shell Diffusion MRI (MDM) dataset. Extensive experimental results show that our CNTSeg-v2 achieves state-of-the-art segmentation performance, outperforming all competing methods.
Anatomy-guided fiber trajectory distribution estimation for cranial nerves tractography
Feb 29, 2024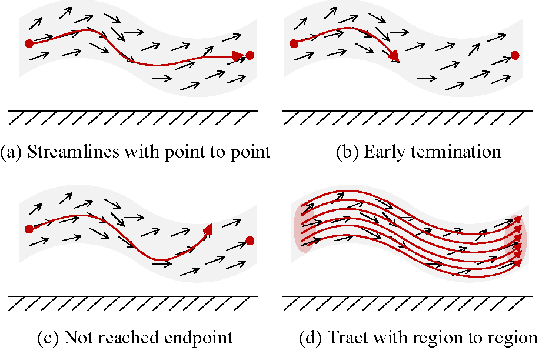
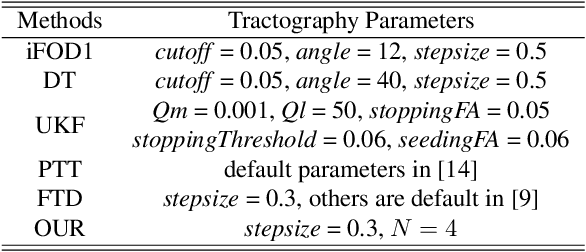
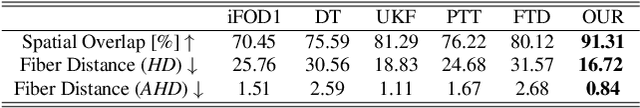
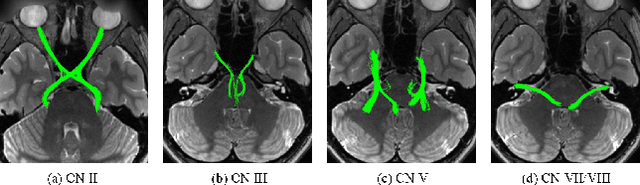
Abstract:Diffusion MRI tractography is an important tool for identifying and analyzing the intracranial course of cranial nerves (CNs). However, the complex environment of the skull base leads to ambiguous spatial correspondence between diffusion directions and fiber geometry, and existing diffusion tractography methods of CNs identification are prone to producing erroneous trajectories and missing true positive connections. To overcome the above challenge, we propose a novel CNs identification framework with anatomy-guided fiber trajectory distribution, which incorporates anatomical shape prior knowledge during the process of CNs tracing to build diffusion tensor vector fields. We introduce higher-order streamline differential equations for continuous flow field representations to directly characterize the fiber trajectory distribution of CNs from the tract-based level. The experimental results on the vivo HCP dataset and the clinical MDM dataset demonstrate that the proposed method reduces false-positive fiber production compared to competing methods and produces reconstructed CNs (i.e. CN II, CN III, CN V, and CN VII/VIII) that are judged to better correspond to the known anatomy.
DeepRGVP: A Novel Microstructure-Informed Supervised Contrastive Learning Framework for Automated Identification Of The Retinogeniculate Pathway Using dMRI Tractography
Nov 15, 2022



Abstract:The retinogeniculate pathway (RGVP) is responsible for carrying visual information from the retina to the lateral geniculate nucleus. Identification and visualization of the RGVP are important in studying the anatomy of the visual system and can inform treatment of related brain diseases. Diffusion MRI (dMRI) tractography is an advanced imaging method that uniquely enables in vivo mapping of the 3D trajectory of the RGVP. Currently, identification of the RGVP from tractography data relies on expert (manual) selection of tractography streamlines, which is time-consuming, has high clinical and expert labor costs, and affected by inter-observer variability. In this paper, we present what we believe is the first deep learning framework, namely DeepRGVP, to enable fast and accurate identification of the RGVP from dMRI tractography data. We design a novel microstructure-informed supervised contrastive learning method that leverages both streamline label and tissue microstructure information to determine positive and negative pairs. We propose a simple and successful streamline-level data augmentation method to address highly imbalanced training data, where the number of RGVP streamlines is much lower than that of non-RGVP streamlines. We perform comparisons with several state-of-the-art deep learning methods that were designed for tractography parcellation, and we show superior RGVP identification results using DeepRGVP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge