Yongjin Zhou
A Study on the Performance of Generative Pre-trained Transformer in Simulating Depressed Individuals on the Standardized Depressive Symptom Scale
Jul 17, 2023



Abstract:Background: Depression is a common mental disorder with societal and economic burden. Current diagnosis relies on self-reports and assessment scales, which have reliability issues. Objective approaches are needed for diagnosing depression. Objective: Evaluate the potential of GPT technology in diagnosing depression. Assess its ability to simulate individuals with depression and investigate the influence of depression scales. Methods: Three depression-related assessment tools (HAMD-17, SDS, GDS-15) were used. Two experiments simulated GPT responses to normal individuals and individuals with depression. Compare GPT's responses with expected results, assess its understanding of depressive symptoms, and performance differences under different conditions. Results: GPT's performance in depression assessment was evaluated. It aligned with scoring criteria for both individuals with depression and normal individuals. Some performance differences were observed based on depression severity. GPT performed better on scales with higher sensitivity. Conclusion: GPT accurately simulates individuals with depression and normal individuals during depression-related assessments. Deviations occur when simulating different degrees of depression, limiting understanding of mild and moderate cases. GPT performs better on scales with higher sensitivity, indicating potential for developing more effective depression scales. GPT has important potential in depression assessment, supporting clinicians and patients.
Rethinking the optimization process for self-supervised model-driven MRI reconstruction
Mar 18, 2022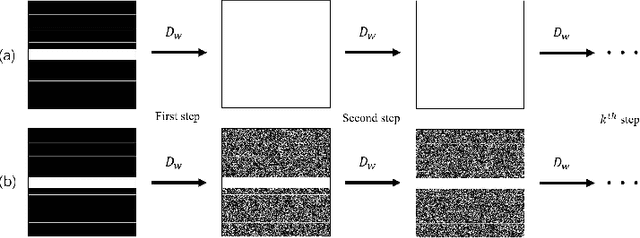
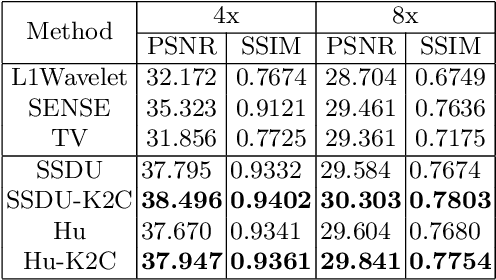
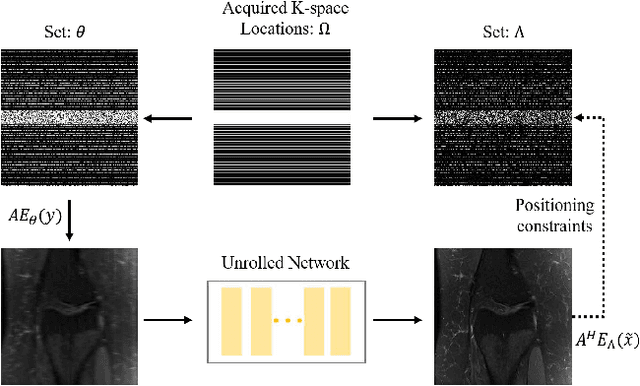

Abstract:Recovering high-quality images from undersampled measurements is critical for accelerated MRI reconstruction. Recently, various supervised deep learning-based MRI reconstruction methods have been developed. Despite the achieved promising performances, these methods require fully sampled reference data, the acquisition of which is resource-intensive and time-consuming. Self-supervised learning has emerged as a promising solution to alleviate the reliance on fully sampled datasets. However, existing self-supervised methods suffer from reconstruction errors due to the insufficient constraint enforced on the non-sampled data points and the error accumulation happened alongside the iterative image reconstruction process for model-driven deep learning reconstrutions. To address these challenges, we propose K2Calibrate, a K-space adaptation strategy for self-supervised model-driven MR reconstruction optimization. By iteratively calibrating the learned measurements, K2Calibrate can reduce the network's reconstruction deterioration caused by statistically dependent noise. Extensive experiments have been conducted on the open-source dataset FastMRI, and K2Calibrate achieves better results than five state-of-the-art methods. The proposed K2Calibrate is plug-and-play and can be easily integrated with different model-driven deep learning reconstruction methods.
D-UNet: a dimension-fusion U shape network for chronic stroke lesion segmentation
Aug 14, 2019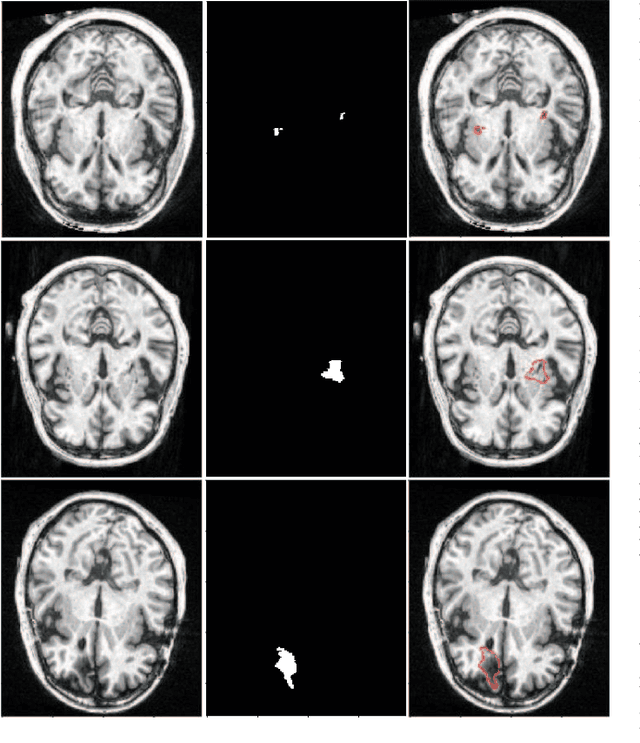
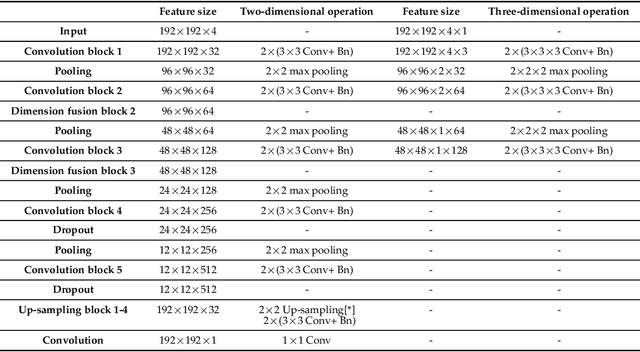
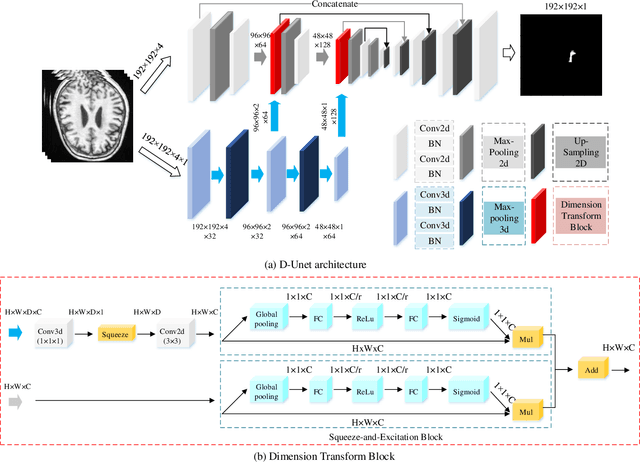
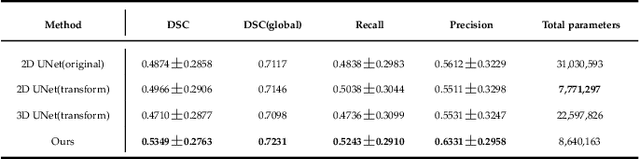
Abstract:Assessing the location and extent of lesions caused by chronic stroke is critical for medical diagnosis, surgical planning, and prognosis. In recent years, with the rapid development of 2D and 3D convolutional neural networks (CNN), the encoder-decoder structure has shown great potential in the field of medical image segmentation. However, the 2D CNN ignores the 3D information of medical images, while the 3D CNN suffers from high computational resource demands. This paper proposes a new architecture called dimension-fusion-UNet (D-UNet), which combines 2D and 3D convolution innovatively in the encoding stage. The proposed architecture achieves a better segmentation performance than 2D networks, while requiring significantly less computation time in comparison to 3D networks. Furthermore, to alleviate the data imbalance issue between positive and negative samples for the network training, we propose a new loss function called Enhance Mixing Loss (EML). This function adds a weighted focal coefficient and combines two traditional loss functions. The proposed method has been tested on the ATLAS dataset and compared to three state-of-the-art methods. The results demonstrate that the proposed method achieves the best quality performance in terms of DSC = 0.5349+0.2763 and precision = 0.6331+0.295).
Classifying Mammographic Breast Density by Residual Learning
Sep 21, 2018



Abstract:Mammographic breast density, a parameter used to describe the proportion of breast tissue fibrosis, is widely adopted as an evaluation characteristic of the likelihood of breast cancer incidence. In this study, we present a radiomics approach based on residual learning for the classification of mammographic breast densities. Our method possesses several encouraging properties such as being almost fully automatic, possessing big model capacity and flexibility. It can obtain outstanding classification results without the necessity of result compensation using mammographs taken from different views. The proposed method was instantiated with the INbreast dataset and classification accuracies of 92.6% and 96.8% were obtained for the four BI-RADS (Breast Imaging and Reporting Data System) category task and the two BI-RADS category task,respectively. The superior performances achieved compared to the existing state-of-the-art methods along with its encouraging properties indicate that our method has a great potential to be applied as a computer-aided diagnosis tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge