Zhizheng Zhuo
Unsupervised Brain Tumor Segmentation with Image-based Prompts
Apr 04, 2023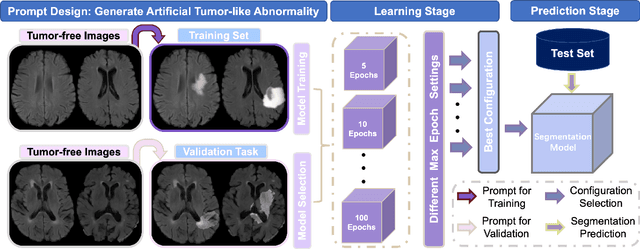
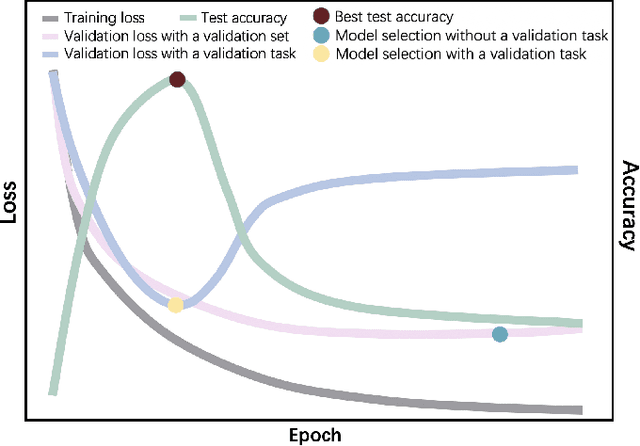
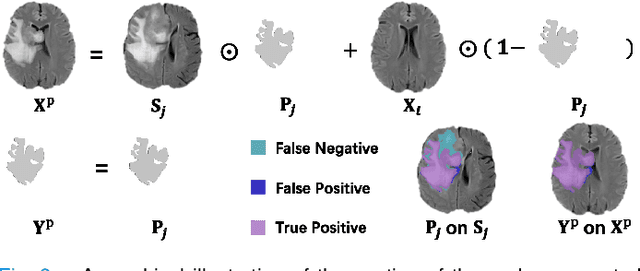
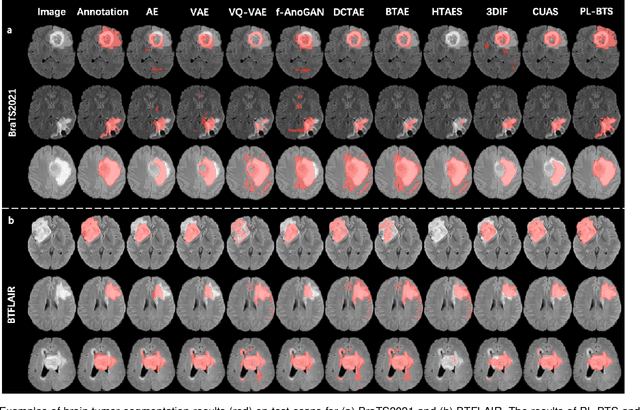
Abstract:Automated brain tumor segmentation based on deep learning (DL) has achieved promising performance. However, it generally relies on annotated images for model training, which is not always feasible in clinical settings. Therefore, the development of unsupervised DL-based brain tumor segmentation approaches without expert annotations is desired. Motivated by the success of prompt learning (PL) in natural language processing, we propose an approach to unsupervised brain tumor segmentation by designing image-based prompts that allow indication of brain tumors, and this approach is dubbed as PL-based Brain Tumor Segmentation (PL-BTS). Specifically, instead of directly training a model for brain tumor segmentation with a large amount of annotated data, we seek to train a model that can answer the question: is a voxel in the input image associated with tumor-like hyper-/hypo-intensity? Such a model can be trained by artificially generating tumor-like hyper-/hypo-intensity on images without tumors with hand-crafted designs. Since the hand-crafted designs may be too simplistic to represent all kinds of real tumors, the trained model may overfit the simplistic hand-crafted task rather than actually answer the question of abnormality. To address this problem, we propose the use of a validation task, where we generate a different hand-crafted task to monitor overfitting. In addition, we propose PL-BTS+ that further improves PL-BTS by exploiting unannotated images with brain tumors. Compared with competing unsupervised methods, the proposed method has achieved marked improvements on both public and in-house datasets, and we have also demonstrated its possible extension to other brain lesion segmentation tasks.
Multiclass Spinal Cord Tumor Segmentation on MRI with Deep Learning
Jan 14, 2021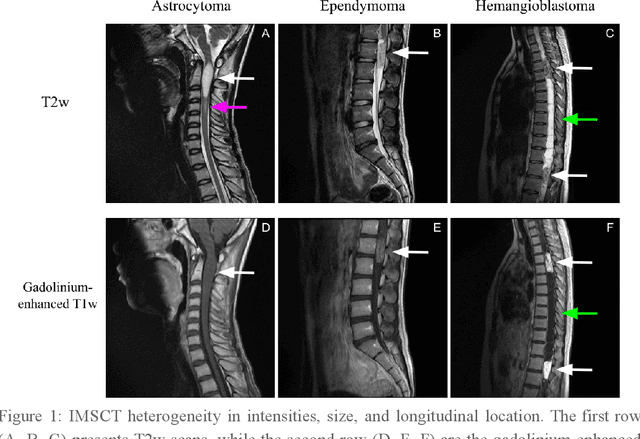
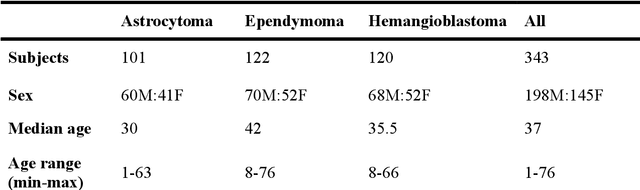
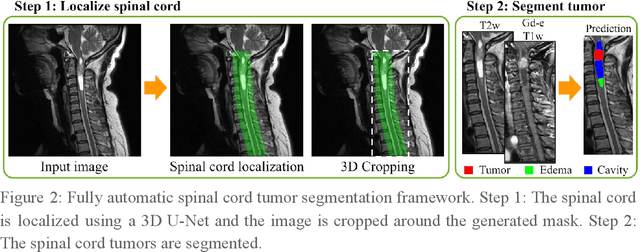
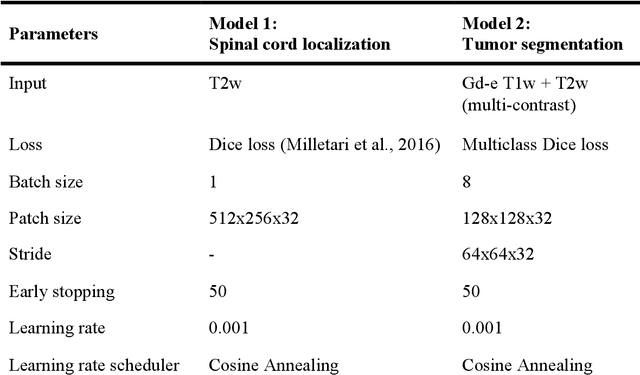
Abstract:Spinal cord tumors lead to neurological morbidity and mortality. Being able to obtain morphometric quantification (size, location, growth rate) of the tumor, edema, and cavity can result in improved monitoring and treatment planning. Such quantification requires the segmentation of these structures into three separate classes. However, manual segmentation of 3-dimensional structures is time-consuming and tedious, motivating the development of automated methods. Here, we tailor a model adapted to the spinal cord tumor segmentation task. Data were obtained from 343 patients using gadolinium-enhanced T1-weighted and T2-weighted MRI scans with cervical, thoracic, and/or lumbar coverage. The dataset includes the three most common intramedullary spinal cord tumor types: astrocytomas, ependymomas, and hemangioblastomas. The proposed approach is a cascaded architecture with U-Net-based models that segments tumors in a two-stage process: locate and label. The model first finds the spinal cord and generates bounding box coordinates. The images are cropped according to this output, leading to a reduced field of view, which mitigates class imbalance. The tumor is then segmented. The segmentation of the tumor, cavity, and edema (as a single class) reached 76.7 $\pm$ 1.5% of Dice score and the segmentation of tumors alone reached 61.8 $\pm$ 4.0% Dice score. The true positive detection rate was above 87% for tumor, edema, and cavity. To the best of our knowledge, this is the first fully automatic deep learning model for spinal cord tumor segmentation. The multiclass segmentation pipeline is available in the Spinal Cord Toolbox (https://spinalcordtoolbox.com/). It can be run with custom data on a regular computer within seconds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge