Olivier Vincent
Impact of individual rater style on deep learning uncertainty in medical imaging segmentation
May 05, 2021



Abstract:While multiple studies have explored the relation between inter-rater variability and deep learning model uncertainty in medical segmentation tasks, little is known about the impact of individual rater style. This study quantifies rater style in the form of bias and consistency and explores their impacts when used to train deep learning models. Two multi-rater public datasets were used, consisting of brain multiple sclerosis lesion and spinal cord grey matter segmentation. On both datasets, results show a correlation ($R^2 = 0.60$ and $0.93$) between rater bias and deep learning uncertainty. The impact of label fusion between raters' annotations on this relationship is also explored, and we show that multi-center consensuses are more effective than single-center consensuses to reduce uncertainty, since rater style is mostly center-specific.
Benefits of Linear Conditioning for Segmentation using Metadata
Feb 18, 2021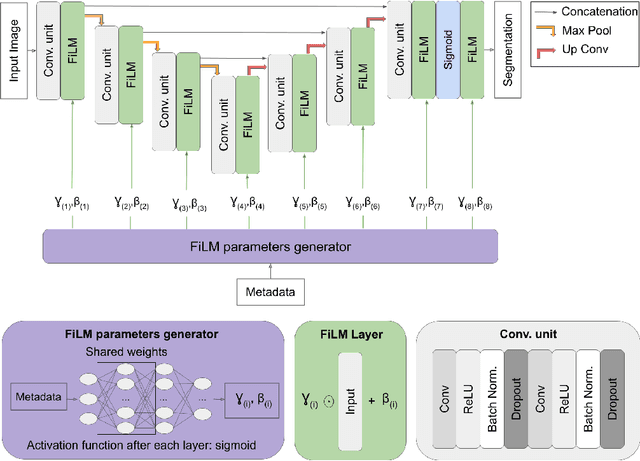

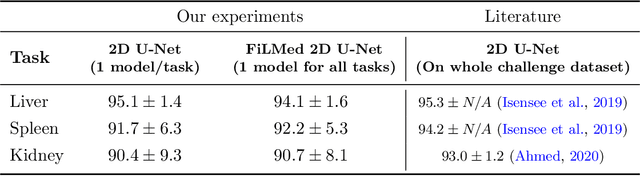

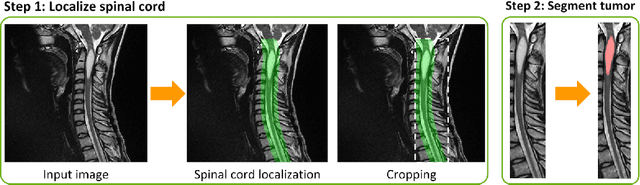
Abstract:Medical images are often accompanied by metadata describing the image (vendor, acquisition parameters) and the patient (disease type or severity, demographics, genomics). This metadata is usually disregarded by image segmentation methods. In this work, we adapt a linear conditioning method called FiLM (Feature-wise Linear Modulation) for image segmentation tasks. This FiLM adaptation enables integrating metadata into segmentation models for better performance. We observed an average Dice score increase of 5.1% on spinal cord tumor segmentation when incorporating the tumor type with FiLM. The metadata modulates the segmentation process through low-cost affine transformations applied on feature maps which can be included in any neural network's architecture. Additionally, we assess the relevance of segmentation FiLM layers for tackling common challenges in medical imaging: training with limited or unbalanced number of annotated data, multi-class training with missing segmentations, and model adaptation to multiple tasks. Our results demonstrated the following benefits of FiLM for segmentation: FiLMed U-Net was robust to missing labels and reached higher Dice scores with few labels (up to 16.7%) compared to single-task U-Net. The code is open-source and available at www.ivadomed.org.
ivadomed: A Medical Imaging Deep Learning Toolbox
Oct 20, 2020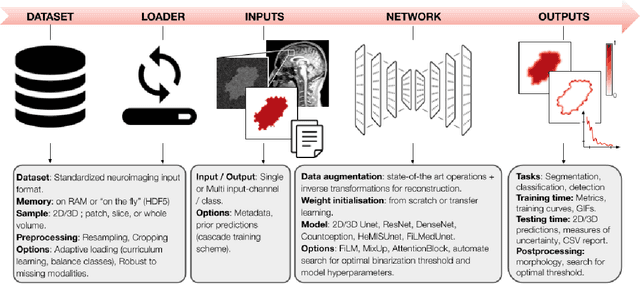
Abstract:ivadomed is an open-source Python package for designing, end-to-end training, and evaluating deep learning models applied to medical imaging data. The package includes APIs, command-line tools, documentation, and tutorials. ivadomed also includes pre-trained models such as spinal tumor segmentation and vertebral labeling. Original features of ivadomed include a data loader that can parse image metadata (e.g., acquisition parameters, image contrast, resolution) and subject metadata (e.g., pathology, age, sex) for custom data splitting or extra information during training and evaluation. Any dataset following the Brain Imaging Data Structure (BIDS) convention will be compatible with ivadomed without the need to manually organize the data, which is typically a tedious task. Beyond the traditional deep learning methods, ivadomed features cutting-edge architectures, such as FiLM and HeMis, as well as various uncertainty estimation methods (aleatoric and epistemic), and losses adapted to imbalanced classes and non-binary predictions. Each step is conveniently configurable via a single file. At the same time, the code is highly modular to allow addition/modification of an architecture or pre/post-processing steps. Example applications of ivadomed include MRI object detection, segmentation, and labeling of anatomical and pathological structures. Overall, ivadomed enables easy and quick exploration of the latest advances in deep learning for medical imaging applications. ivadomed's main project page is available at https://ivadomed.org.
Automatic segmentation of spinal multiple sclerosis lesions: How to generalize across MRI contrasts?
Mar 11, 2020

Abstract:Despite recent improvements in medical image segmentation, the ability to generalize across imaging contrasts remains an open issue. To tackle this challenge, we implement Feature-wise Linear Modulation (FiLM) to leverage physics knowledge within the segmentation model and learn the characteristics of each contrast. Interestingly, a well-optimised U-Net reached the same performance as our FiLMed-Unet on a multi-contrast dataset (0.72 of Dice score), which suggests that there is a bottleneck in spinal MS lesion segmentation different from the generalization across varying contrasts. This bottleneck likely stems from inter-rater variability, which is estimated at 0.61 of Dice score in our dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge