Kim-Han Thung
Brain Tissue Segmentation Across the Human Lifespan via Supervised Contrastive Learning
Jan 03, 2023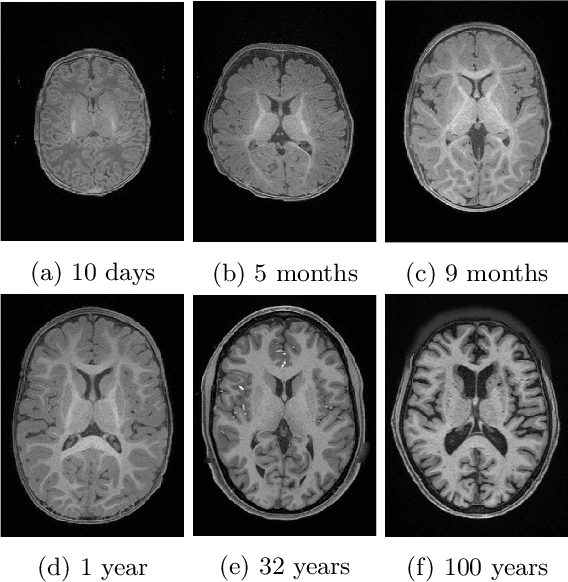
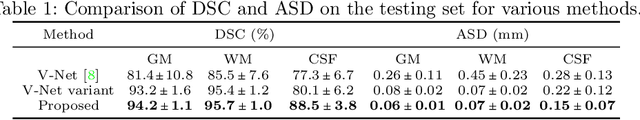
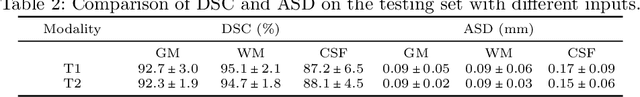
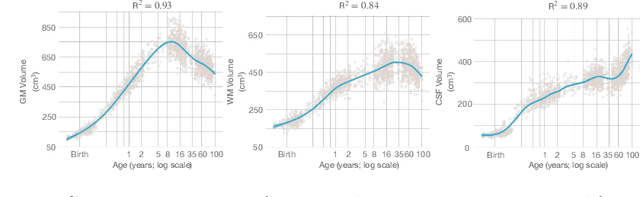
Abstract:Automatic segmentation of brain MR images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) is critical for tissue volumetric analysis and cortical surface reconstruction. Due to dramatic structural and appearance changes associated with developmental and aging processes, existing brain tissue segmentation methods are only viable for specific age groups. Consequently, methods developed for one age group may fail for another. In this paper, we make the first attempt to segment brain tissues across the entire human lifespan (0-100 years of age) using a unified deep learning model. To overcome the challenges related to structural variability underpinned by biological processes, intensity inhomogeneity, motion artifacts, scanner-induced differences, and acquisition protocols, we propose to use contrastive learning to improve the quality of feature representations in a latent space for effective lifespan tissue segmentation. We compared our approach with commonly used segmentation methods on a large-scale dataset of 2,464 MR images. Experimental results show that our model accurately segments brain tissues across the lifespan and outperforms existing methods.
Learning MRI Artifact Removal With Unpaired Data
Oct 09, 2021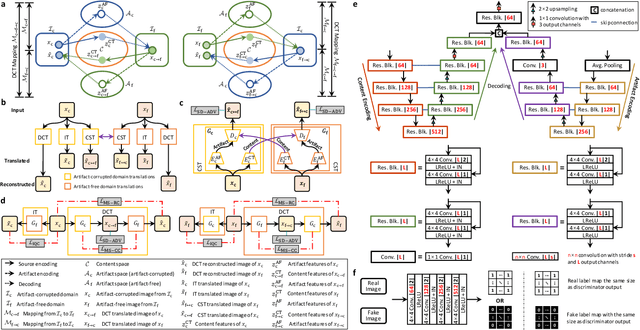
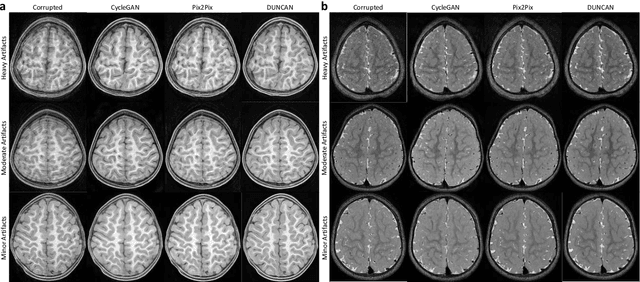
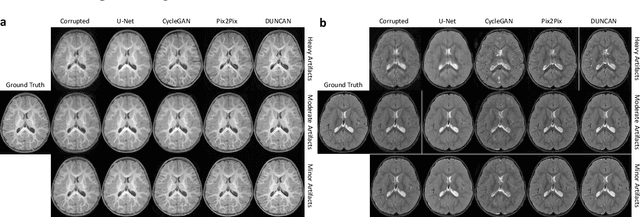
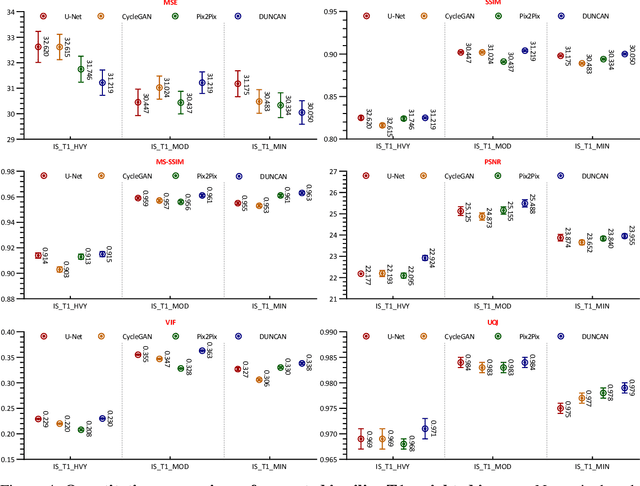
Abstract:Retrospective artifact correction (RAC) improves image quality post acquisition and enhances image usability. Recent machine learning driven techniques for RAC are predominantly based on supervised learning and therefore practical utility can be limited as data with paired artifact-free and artifact-corrupted images are typically insufficient or even non-existent. Here we show that unwanted image artifacts can be disentangled and removed from an image via an RAC neural network learned with unpaired data. This implies that our method does not require matching artifact-corrupted data to be either collected via acquisition or generated via simulation. Experimental results demonstrate that our method is remarkably effective in removing artifacts and retaining anatomical details in images with different contrasts.
Learning-based Computer-aided Prescription Model for Parkinson's Disease: A Data-driven Perspective
Jul 31, 2020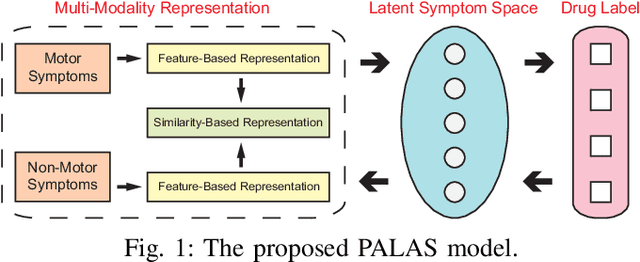
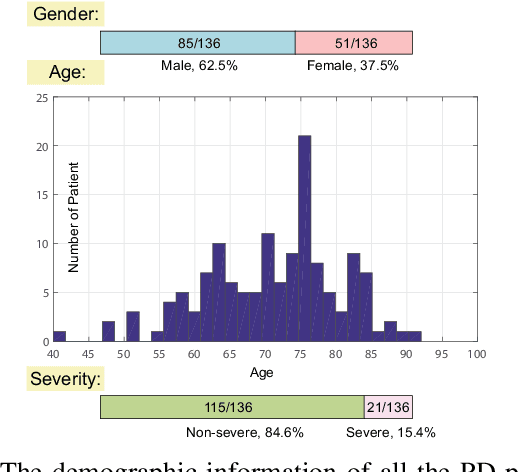
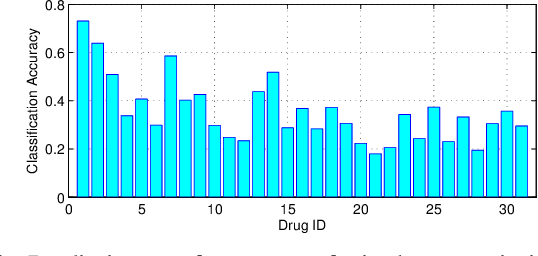
Abstract:In this paper, we study a novel problem: "automatic prescription recommendation for PD patients." To realize this goal, we first build a dataset by collecting 1) symptoms of PD patients, and 2) their prescription drug provided by neurologists. Then, we build a novel computer-aided prescription model by learning the relation between observed symptoms and prescription drug. Finally, for the new coming patients, we could recommend (predict) suitable prescription drug on their observed symptoms by our prescription model. From the methodology part, our proposed model, namely Prescription viA Learning lAtent Symptoms (PALAS), could recommend prescription using the multi-modality representation of the data. In PALAS, a latent symptom space is learned to better model the relationship between symptoms and prescription drug, as there is a large semantic gap between them. Moreover, we present an efficient alternating optimization method for PALAS. We evaluated our method using the data collected from 136 PD patients at Nanjing Brain Hospital, which can be regarded as a large dataset in PD research community. The experimental results demonstrate the effectiveness and clinical potential of our method in this recommendation task, if compared with other competing methods.
Real-Time Quality Assessment of Pediatric MRI via Semi-Supervised Deep Nonlocal Residual Neural Networks
Apr 07, 2019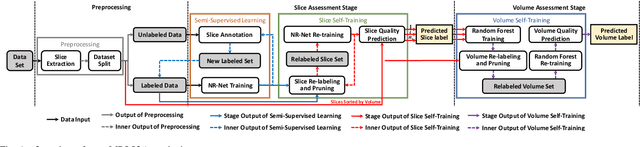
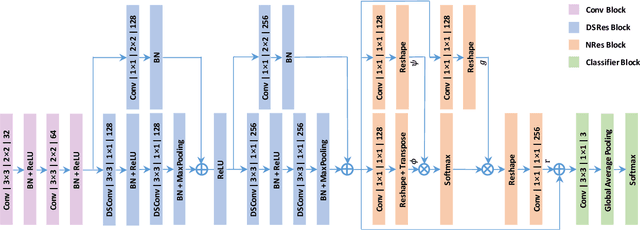
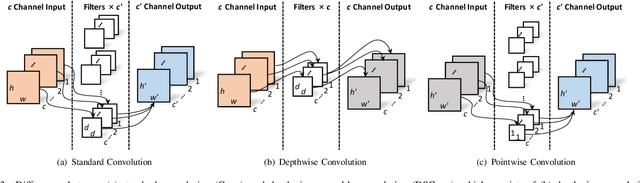
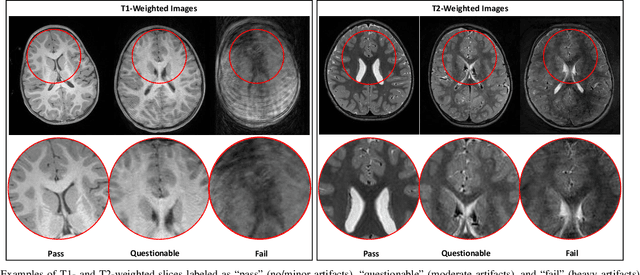
Abstract:In this paper, we introduce an image quality assessment (IQA) method for pediatric T1- and T2-weighted MR images. IQA is first performed slice-wise using a nonlocal residual neural network (NR-Net) and then volume-wise by agglomerating the slice QA results using random forest. Our method requires only a small amount of quality-annotated images for training and is designed to be robust to annotation noise that might occur due to rater errors and the inevitable mix of good and bad slices in an image volume. Using a small set of quality-assessed images, we pre-train NR-Net to annotate each image slice with an initial quality rating (i.e., pass, questionable, fail), which we then refine by semi-supervised learning and iterative self-training. Experimental results demonstrate that our method, trained using only samples of modest size, exhibit great generalizability, capable of real-time (milliseconds per volume) large-scale IQA with near-perfect accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge