Hao Guan
School of Computer Science, The University of Sydney
VTC-R1: Vision-Text Compression for Efficient Long-Context Reasoning
Jan 29, 2026Abstract:Long-context reasoning has significantly empowered large language models (LLMs) to tackle complex tasks, yet it introduces severe efficiency bottlenecks due to the computational complexity. Existing efficient approaches often rely on complex additional training or external models for compression, which limits scalability and discards critical fine-grained information. In this paper, we propose VTC-R1, a new efficient reasoning paradigm that integrates vision-text compression into the reasoning process. Instead of processing lengthy textual traces, VTC-R1 renders intermediate reasoning segments into compact images, which are iteratively fed back into vision-language models as "optical memory." We construct a training dataset based on OpenR1-Math-220K achieving 3.4x token compression and fine-tune representative VLMs-Glyph and Qwen3-VL. Extensive experiments on benchmarks such as MATH500, AIME25, AMC23 and GPQA-D demonstrate that VTC-R1 consistently outperforms standard long-context reasoning. Furthermore, our approach significantly improves inference efficiency, achieving 2.7x speedup in end-to-end latency, highlighting its potential as a scalable solution for reasoning-intensive applications. Our code is available at https://github.com/w-yibo/VTC-R1.
Hot-Start from Pixels: Low-Resolution Visual Tokens for Chinese Language Modeling
Jan 15, 2026Abstract:Large language models typically represent Chinese characters as discrete index-based tokens, largely ignoring their visual form. For logographic scripts, visual structure carries semantic and phonetic information, which may aid prediction. We investigate whether low-resolution visual inputs can serve as an alternative for character-level modeling. Instead of token IDs, our decoder receives grayscale images of individual characters, with resolutions as low as 8 x 8 pixels. Remarkably, these inputs achieve 39.2% accuracy, comparable to the index-based baseline of 39.1%. Such low-resource settings also exhibit a pronounced hot-start effect: by 0.4% of total training, accuracy reaches above 12%, while index-based models lag at below 6%. Overall, our results demonstrate that minimal visual structure can provide a robust and efficient signal for Chinese language modeling, offering an alternative perspective on character representation that complements traditional index-based approaches.
Detecting Performance Degradation under Data Shift in Pathology Vision-Language Model
Jan 02, 2026Abstract:Vision-Language Models have demonstrated strong potential in medical image analysis and disease diagnosis. However, after deployment, their performance may deteriorate when the input data distribution shifts from that observed during development. Detecting such performance degradation is essential for clinical reliability, yet remains challenging for large pre-trained VLMs operating without labeled data. In this study, we investigate performance degradation detection under data shift in a state-of-the-art pathology VLM. We examine both input-level data shift and output-level prediction behavior to understand their respective roles in monitoring model reliability. To facilitate systematic analysis of input data shift, we develop DomainSAT, a lightweight toolbox with a graphical interface that integrates representative shift detection algorithms and enables intuitive exploration of data shift. Our analysis shows that while input data shift detection is effective at identifying distributional changes and providing early diagnostic signals, it does not always correspond to actual performance degradation. Motivated by this observation, we further study output-based monitoring and introduce a label-free, confidence-based degradation indicator that directly captures changes in model prediction confidence. We find that this indicator exhibits a close relationship with performance degradation and serves as an effective complement to input shift detection. Experiments on a large-scale pathology dataset for tumor classification demonstrate that combining input data shift detection and output confidence-based indicators enables more reliable detection and interpretation of performance degradation in VLMs under data shift. These findings provide a practical and complementary framework for monitoring the reliability of foundation models in digital pathology.
SINKT: A Structure-Aware Inductive Knowledge Tracing Model with Large Language Model
Jul 01, 2024



Abstract:Knowledge Tracing (KT) aims to determine whether students will respond correctly to the next question, which is a crucial task in intelligent tutoring systems (ITS). In educational KT scenarios, transductive ID-based methods often face severe data sparsity and cold start problems, where interactions between individual students and questions are sparse, and new questions and concepts consistently arrive in the database. In addition, existing KT models only implicitly consider the correlation between concepts and questions, lacking direct modeling of the more complex relationships in the heterogeneous graph of concepts and questions. In this paper, we propose a Structure-aware Inductive Knowledge Tracing model with large language model (dubbed SINKT), which, for the first time, introduces large language models (LLMs) and realizes inductive knowledge tracing. Firstly, SINKT utilizes LLMs to introduce structural relationships between concepts and constructs a heterogeneous graph for concepts and questions. Secondly, by encoding concepts and questions with LLMs, SINKT incorporates semantic information to aid prediction. Finally, SINKT predicts the student's response to the target question by interacting with the student's knowledge state and the question representation. Experiments on four real-world datasets demonstrate that SINKT achieves state-of-the-art performance among 12 existing transductive KT models. Additionally, we explore the performance of SINKT on the inductive KT task and provide insights into various modules.
PAODING: A High-fidelity Data-free Pruning Toolkit for Debloating Pre-trained Neural Networks
Apr 30, 2024

Abstract:We present PAODING, a toolkit to debloat pretrained neural network models through the lens of data-free pruning. To preserve the model fidelity, PAODING adopts an iterative process, which dynamically measures the effect of deleting a neuron to identify candidates that have the least impact to the output layer. Our evaluation shows that PAODING can significantly reduce the model size, generalize on different datasets and models, and meanwhile preserve the model fidelity in terms of test accuracy and adversarial robustness. PAODING is publicly available on PyPI via https://pypi.org/project/paoding-dl.
Federated Learning for Medical Image Analysis: A Survey
Jun 16, 2023Abstract:Machine learning in medical imaging often faces a fundamental dilemma, namely the small sample size problem. Many recent studies suggest using multi-domain data pooled from different acquisition sites/datasets to improve statistical power. However, medical images from different sites cannot be easily shared to build large datasets for model training due to privacy protection reasons. As a promising solution, federated learning, which enables collaborative training of machine learning models based on data from different sites without cross-site data sharing, has attracted considerable attention recently. In this paper, we conduct a comprehensive survey of the recent development of federated learning methods in medical image analysis. We first introduce the background and motivation of federated learning for dealing with privacy protection and collaborative learning issues in medical imaging. We then present a comprehensive review of recent advances in federated learning methods for medical image analysis. Specifically, existing methods are categorized based on three critical aspects of a federated learning system, including client end, server end, and communication techniques. In each category, we summarize the existing federated learning methods according to specific research problems in medical image analysis and also provide insights into the motivations of different approaches. In addition, we provide a review of existing benchmark medical imaging datasets and software platforms for current federated learning research. We also conduct an experimental study to empirically evaluate typical federated learning methods for medical image analysis. This survey can help to better understand the current research status, challenges and potential research opportunities in this promising research field.
DomainATM: Domain Adaptation Toolbox for Medical Data Analysis
Sep 24, 2022



Abstract:Domain adaptation (DA) is an important technique for modern machine learning-based medical data analysis, which aims at reducing distribution differences between different medical datasets. A proper domain adaptation method can significantly enhance the statistical power by pooling data acquired from multiple sites/centers. To this end, we have developed the Domain Adaptation Toolbox for Medical data analysis (DomainATM) - an open-source software package designed for fast facilitation and easy customization of domain adaptation methods for medical data analysis. The DomainATM is implemented in MATLAB with a user-friendly graphical interface, and it consists of a collection of popular data adaptation algorithms that have been extensively applied to medical image analysis and computer vision. With DomainATM, researchers are able to facilitate fast feature-level and image-level adaptation, visualization and performance evaluation of different adaptation methods for medical data analysis. More importantly, the DomainATM enables the users to develop and test their own adaptation methods through scripting, greatly enhancing its utility and extensibility. An overview characteristic and usage of DomainATM is presented and illustrated with three example experiments, demonstrating its effectiveness, simplicity, and flexibility. The software, source code, and manual are available online.
Attention-Guided Autoencoder for Automated Progression Prediction of Subjective Cognitive Decline with Structural MRI
Jun 24, 2022



Abstract:Subjective cognitive decline (SCD) is a preclinical stage of Alzheimer's disease (AD) which occurs even before mild cognitive impairment (MCI). Progressive SCD will convert to MCI with the potential of further evolving to AD. Therefore, early identification of progressive SCD with neuroimaging techniques (e.g., structural MRI) is of great clinical value for early intervention of AD. However, existing MRI-based machine/deep learning methods usually suffer the small-sample-size problem which poses a great challenge to related neuroimaging analysis. The central question we aim to tackle in this paper is how to leverage related domains (e.g., AD/NC) to assist the progression prediction of SCD. Meanwhile, we are concerned about which brain areas are more closely linked to the identification of progressive SCD. To this end, we propose an attention-guided autoencoder model for efficient cross-domain adaptation which facilitates the knowledge transfer from AD to SCD. The proposed model is composed of four key components: 1) a feature encoding module for learning shared subspace representations of different domains, 2) an attention module for automatically locating discriminative brain regions of interest defined in brain atlases, 3) a decoding module for reconstructing the original input, 4) a classification module for identification of brain diseases. Through joint training of these four modules, domain invariant features can be learned. Meanwhile, the brain disease related regions can be highlighted by the attention mechanism. Extensive experiments on the publicly available ADNI dataset and a private CLAS dataset have demonstrated the effectiveness of the proposed method. The proposed model is straightforward to train and test with only 5-10 seconds on CPUs and is suitable for medical tasks with small datasets.
Brain Age Estimation From MRI Using Cascade Networks with Ranking Loss
Jun 06, 2021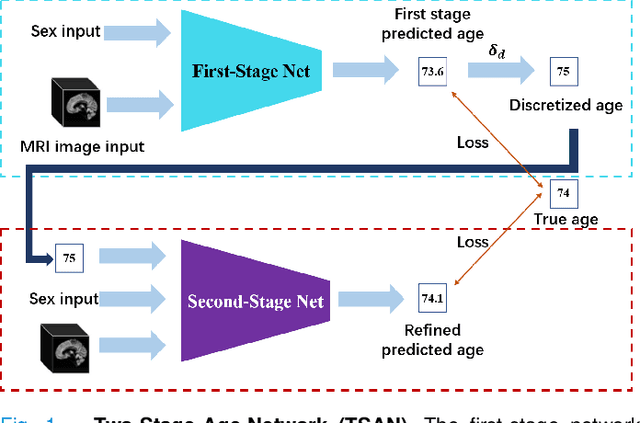
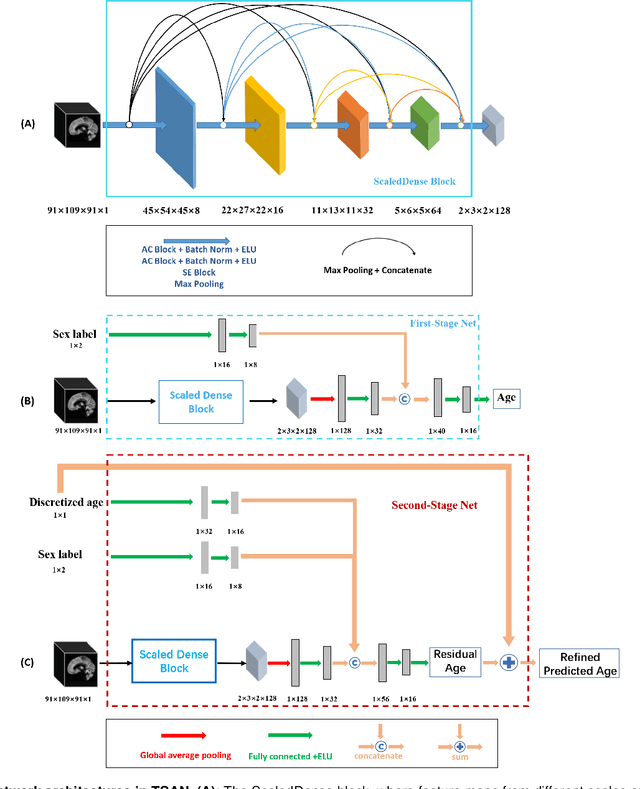
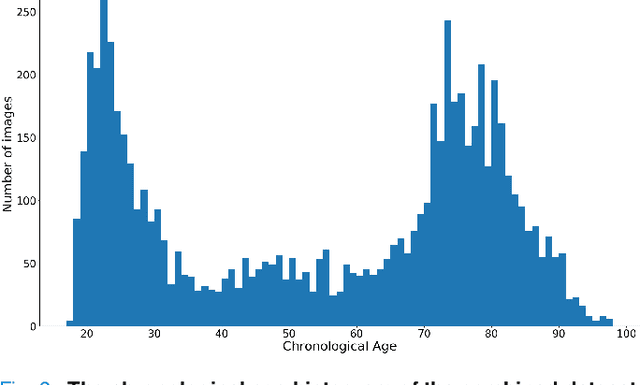
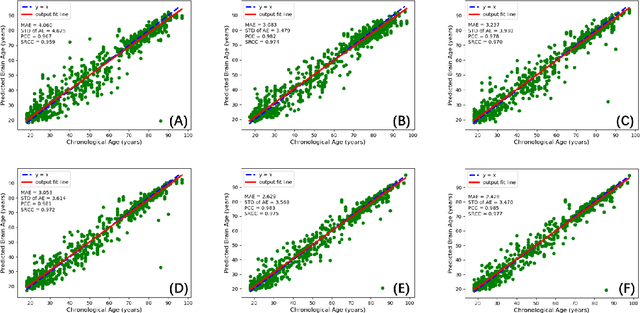
Abstract:Chronological age of healthy people is able to be predicted accurately using deep neural networks from neuroimaging data, and the predicted brain age could serve as a biomarker for detecting aging-related diseases. In this paper, a novel 3D convolutional network, called two-stage-age-network (TSAN), is proposed to estimate brain age from T1-weighted MRI data. Compared with existing methods, TSAN has the following improvements. First, TSAN uses a two-stage cascade network architecture, where the first-stage network estimates a rough brain age, then the second-stage network estimates the brain age more accurately from the discretized brain age by the first-stage network. Second, to our knowledge, TSAN is the first work to apply novel ranking losses in brain age estimation, together with the traditional mean square error (MSE) loss. Third, densely connected paths are used to combine feature maps with different scales. The experiments with $6586$ MRIs showed that TSAN could provide accurate brain age estimation, yielding mean absolute error (MAE) of $2.428$ and Pearson's correlation coefficient (PCC) of $0.985$, between the estimated and chronological ages. Furthermore, using the brain age gap between brain age and chronological age as a biomarker, Alzheimer's disease (AD) and Mild Cognitive Impairment (MCI) can be distinguished from healthy control (HC) subjects by support vector machine (SVM). Classification AUC in AD/HC and MCI/HC was $0.904$ and $0.823$, respectively. It showed that brain age gap is an effective biomarker associated with risk of dementia, and has potential for early-stage dementia risk screening. The codes and trained models have been released on GitHub: https://github.com/Milan-BUAA/TSAN-brain-age-estimation.
MRI-based Alzheimer's disease prediction via distilling the knowledge in multi-modal data
Apr 08, 2021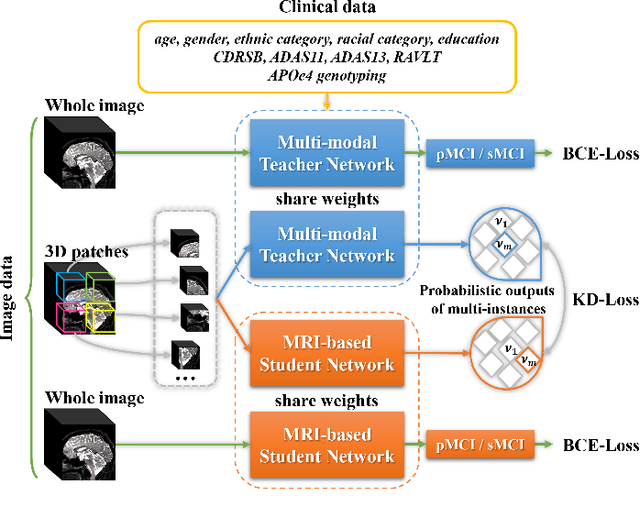
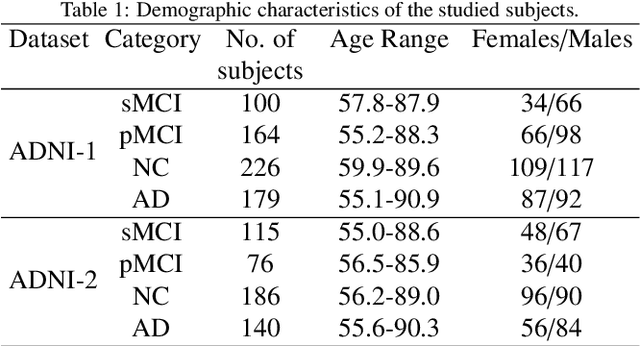
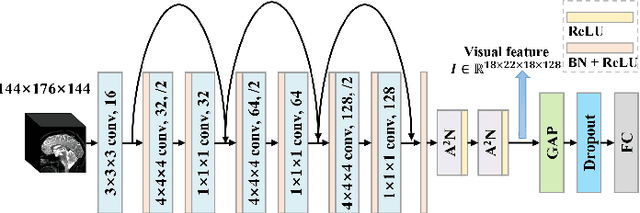
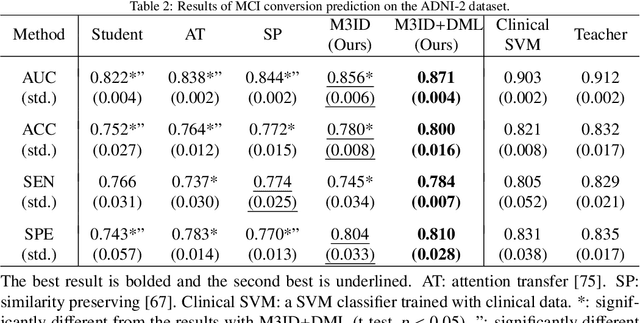
Abstract:Mild cognitive impairment (MCI) conversion prediction, i.e., identifying MCI patients of high risks converting to Alzheimer's disease (AD), is essential for preventing or slowing the progression of AD. Although previous studies have shown that the fusion of multi-modal data can effectively improve the prediction accuracy, their applications are largely restricted by the limited availability or high cost of multi-modal data. Building an effective prediction model using only magnetic resonance imaging (MRI) remains a challenging research topic. In this work, we propose a multi-modal multi-instance distillation scheme, which aims to distill the knowledge learned from multi-modal data to an MRI-based network for MCI conversion prediction. In contrast to existing distillation algorithms, the proposed multi-instance probabilities demonstrate a superior capability of representing the complicated atrophy distributions, and can guide the MRI-based network to better explore the input MRI. To our best knowledge, this is the first study that attempts to improve an MRI-based prediction model by leveraging extra supervision distilled from multi-modal information. Experiments demonstrate the advantage of our framework, suggesting its potentials in the data-limited clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge