Gregory Slabaugh
School of Electronic Engineering and Computer Science, Queen Mary University of London, UK, Queen Mary Digital Environment Research Institute
OpenUS: A Fully Open-Source Foundation Model for Ultrasound Image Analysis via Self-Adaptive Masked Contrastive Learning
Nov 14, 2025Abstract:Ultrasound (US) is one of the most widely used medical imaging modalities, thanks to its low cost, portability, real-time feedback, and absence of ionizing radiation. However, US image interpretation remains highly operator-dependent and varies significantly across anatomical regions, acquisition protocols, and device types. These variations, along with unique challenges such as speckle, low contrast, and limited standardized annotations, hinder the development of generalizable, label-efficient ultrasound AI models. In this paper, we propose OpenUS, the first reproducible, open-source ultrasound foundation model built on a large collection of public data. OpenUS employs a vision Mamba backbone, capturing both local and global long-range dependencies across the image. To extract rich features during pre-training, we introduce a novel self-adaptive masking framework that combines contrastive learning with masked image modeling. This strategy integrates the teacher's attention map with student reconstruction loss, adaptively refining clinically-relevant masking to enhance pre-training effectiveness. OpenUS also applies a dynamic learning schedule to progressively adjust the difficulty of the pre-training process. To develop the foundation model, we compile the largest to-date public ultrasound dataset comprising over 308K images from 42 publicly available datasets, covering diverse anatomical regions, institutions, imaging devices, and disease types. Our pre-trained OpenUS model can be easily adapted to specific downstream tasks by serving as a backbone for label-efficient fine-tuning. Code is available at https://github.com/XZheng0427/OpenUS.
ViDAR: Video Diffusion-Aware 4D Reconstruction From Monocular Inputs
Jun 23, 2025Abstract:Dynamic Novel View Synthesis aims to generate photorealistic views of moving subjects from arbitrary viewpoints. This task is particularly challenging when relying on monocular video, where disentangling structure from motion is ill-posed and supervision is scarce. We introduce Video Diffusion-Aware Reconstruction (ViDAR), a novel 4D reconstruction framework that leverages personalised diffusion models to synthesise a pseudo multi-view supervision signal for training a Gaussian splatting representation. By conditioning on scene-specific features, ViDAR recovers fine-grained appearance details while mitigating artefacts introduced by monocular ambiguity. To address the spatio-temporal inconsistency of diffusion-based supervision, we propose a diffusion-aware loss function and a camera pose optimisation strategy that aligns synthetic views with the underlying scene geometry. Experiments on DyCheck, a challenging benchmark with extreme viewpoint variation, show that ViDAR outperforms all state-of-the-art baselines in visual quality and geometric consistency. We further highlight ViDAR's strong improvement over baselines on dynamic regions and provide a new benchmark to compare performance in reconstructing motion-rich parts of the scene. Project page: https://vidar-4d.github.io
DanceChat: Large Language Model-Guided Music-to-Dance Generation
Jun 12, 2025Abstract:Music-to-dance generation aims to synthesize human dance motion conditioned on musical input. Despite recent progress, significant challenges remain due to the semantic gap between music and dance motion, as music offers only abstract cues, such as melody, groove, and emotion, without explicitly specifying the physical movements. Moreover, a single piece of music can produce multiple plausible dance interpretations. This one-to-many mapping demands additional guidance, as music alone provides limited information for generating diverse dance movements. The challenge is further amplified by the scarcity of paired music and dance data, which restricts the model\^a\u{A}\'Zs ability to learn diverse dance patterns. In this paper, we introduce DanceChat, a Large Language Model (LLM)-guided music-to-dance generation approach. We use an LLM as a choreographer that provides textual motion instructions, offering explicit, high-level guidance for dance generation. This approach goes beyond implicit learning from music alone, enabling the model to generate dance that is both more diverse and better aligned with musical styles. Our approach consists of three components: (1) an LLM-based pseudo instruction generation module that produces textual dance guidance based on music style and structure, (2) a multi-modal feature extraction and fusion module that integrates music, rhythm, and textual guidance into a shared representation, and (3) a diffusion-based motion synthesis module together with a multi-modal alignment loss, which ensures that the generated dance is aligned with both musical and textual cues. Extensive experiments on AIST++ and human evaluations show that DanceChat outperforms state-of-the-art methods both qualitatively and quantitatively.
STaR: Seamless Spatial-Temporal Aware Motion Retargeting with Penetration and Consistency Constraints
Apr 09, 2025Abstract:Motion retargeting seeks to faithfully replicate the spatio-temporal motion characteristics of a source character onto a target character with a different body shape. Apart from motion semantics preservation, ensuring geometric plausibility and maintaining temporal consistency are also crucial for effective motion retargeting. However, many existing methods prioritize either geometric plausibility or temporal consistency. Neglecting geometric plausibility results in interpenetration while neglecting temporal consistency leads to motion jitter. In this paper, we propose a novel sequence-to-sequence model for seamless Spatial-Temporal aware motion Retargeting (STaR), with penetration and consistency constraints. STaR consists of two modules: (1) a spatial module that incorporates dense shape representation and a novel limb penetration constraint to ensure geometric plausibility while preserving motion semantics, and (2) a temporal module that utilizes a temporal transformer and a novel temporal consistency constraint to predict the entire motion sequence at once while enforcing multi-level trajectory smoothness. The seamless combination of the two modules helps us achieve a good balance between the semantic, geometric, and temporal targets. Extensive experiments on the Mixamo and ScanRet datasets demonstrate that our method produces plausible and coherent motions while significantly reducing interpenetration rates compared with other approaches.
BioX-CPath: Biologically-driven Explainable Diagnostics for Multistain IHC Computational Pathology
Mar 26, 2025



Abstract:The development of biologically interpretable and explainable models remains a key challenge in computational pathology, particularly for multistain immunohistochemistry (IHC) analysis. We present BioX-CPath, an explainable graph neural network architecture for whole slide image (WSI) classification that leverages both spatial and semantic features across multiple stains. At its core, BioX-CPath introduces a novel Stain-Aware Attention Pooling (SAAP) module that generates biologically meaningful, stain-aware patient embeddings. Our approach achieves state-of-the-art performance on both Rheumatoid Arthritis and Sjogren's Disease multistain datasets. Beyond performance metrics, BioX-CPath provides interpretable insights through stain attention scores, entropy measures, and stain interaction scores, that permit measuring model alignment with known pathological mechanisms. This biological grounding, combined with strong classification performance, makes BioX-CPath particularly suitable for clinical applications where interpretability is key. Source code and documentation can be found at: https://github.com/AmayaGS/BioX-CPath.
HandSplat: Embedding-Driven Gaussian Splatting for High-Fidelity Hand Rendering
Mar 18, 2025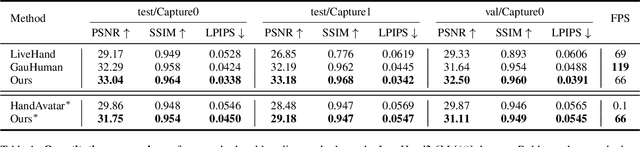
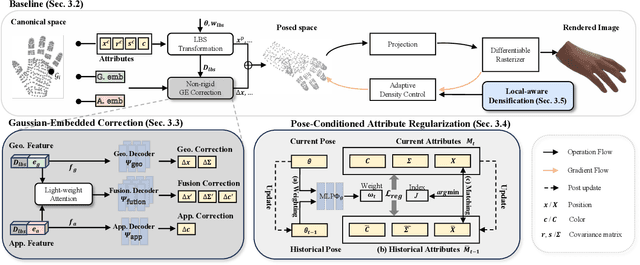
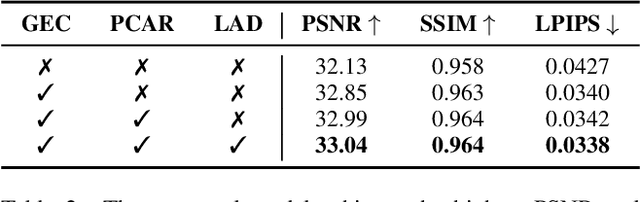

Abstract:Existing 3D Gaussian Splatting (3DGS) methods for hand rendering rely on rigid skeletal motion with an oversimplified non-rigid motion model, which fails to capture fine geometric and appearance details. Additionally, they perform densification based solely on per-point gradients and process poses independently, ignoring spatial and temporal correlations. These limitations lead to geometric detail loss, temporal instability, and inefficient point distribution. To address these issues, we propose HandSplat, a novel Gaussian Splatting-based framework that enhances both fidelity and stability for hand rendering. To improve fidelity, we extend standard 3DGS attributes with implicit geometry and appearance embeddings for finer non-rigid motion modeling while preserving the static hand characteristic modeled by original 3DGS attributes. Additionally, we introduce a local gradient-aware densification strategy that dynamically refines Gaussian density in high-variation regions. To improve stability, we incorporate pose-conditioned attribute regularization to encourage attribute consistency across similar poses, mitigating temporal artifacts. Extensive experiments on InterHand2.6M demonstrate that HandSplat surpasses existing methods in fidelity and stability while achieving real-time performance. We will release the code and pre-trained models upon acceptance.
SuperCap: Multi-resolution Superpixel-based Image Captioning
Mar 11, 2025Abstract:It has been a longstanding goal within image captioning to move beyond a dependence on object detection. We investigate using superpixels coupled with Vision Language Models (VLMs) to bridge the gap between detector-based captioning architectures and those that solely pretrain on large datasets. Our novel superpixel approach ensures that the model receives object-like features whilst the use of VLMs provides our model with open set object understanding. Furthermore, we extend our architecture to make use of multi-resolution inputs, allowing our model to view images in different levels of detail, and use an attention mechanism to determine which parts are most relevant to the caption. We demonstrate our model's performance with multiple VLMs and through a range of ablations detailing the impact of different architectural choices. Our full model achieves a competitive CIDEr score of $136.9$ on the COCO Karpathy split.
HanDrawer: Leveraging Spatial Information to Render Realistic Hands Using a Conditional Diffusion Model in Single Stage
Mar 03, 2025



Abstract:Although diffusion methods excel in text-to-image generation, generating accurate hand gestures remains a major challenge, resulting in severe artifacts, such as incorrect number of fingers or unnatural gestures. To enable the diffusion model to learn spatial information to improve the quality of the hands generated, we propose HanDrawer, a module to condition the hand generation process. Specifically, we apply graph convolutional layers to extract the endogenous spatial structure and physical constraints implicit in MANO hand mesh vertices. We then align and fuse these spatial features with other modalities via cross-attention. The spatially fused features are used to guide a single stage diffusion model denoising process for high quality generation of the hand region. To improve the accuracy of spatial feature fusion, we propose a Position-Preserving Zero Padding (PPZP) fusion strategy, which ensures that the features extracted by HanDrawer are fused into the region of interest in the relevant layers of the diffusion model. HanDrawer learns the entire image features while paying special attention to the hand region thanks to an additional hand reconstruction loss combined with the denoising loss. To accurately train and evaluate our approach, we perform careful cleansing and relabeling of the widely used HaGRID hand gesture dataset and obtain high quality multimodal data. Quantitative and qualitative analyses demonstrate the state-of-the-art performance of our method on the HaGRID dataset through multiple evaluation metrics. Source code and our enhanced dataset will be released publicly if the paper is accepted.
Multimodal Outer Arithmetic Block Dual Fusion of Whole Slide Images and Omics Data for Precision Oncology
Nov 26, 2024Abstract:Developing a central nervous system (CNS) tumor classifier by integrating DNA methylation data with Whole Slide Images (WSI) offers significant potential for enhancing diagnostic precision in neuropathology. Existing approaches typically integrate encoded omic data with histology only once - either at an early or late fusion stage - while reintroducing encoded omic data to create a dual fusion variant remains unexplored. Nevertheless, reintroduction of omic embeddings during early and late fusion enables the capture of complementary information from localized patch-level and holistic slide-level interactions, allowing boosted performance through advanced multimodal integration. To achieve this, we propose a dual fusion framework that integrates omic data at both early and late stages, fully leveraging its diagnostic strength. In the early fusion stage, omic embeddings are projected into a patch-wise latent space, generating omic-WSI embeddings that encapsulate per-patch molecular and morphological insights, effectively incorporating this information into the spatial representation of histology. These embeddings are refined with a multiple instance learning gated attention mechanism to attend to critical patches. In the late fusion stage, we reintroduce the omic data by fusing it with slide-level omic-WSI embeddings using a Multimodal Outer Arithmetic Block (MOAB), which richly intermingles features from both modalities, capturing their global correlations and complementarity. We demonstrate accurate CNS tumor subtyping across 20 fine-grained subtypes and validate our approach on benchmark datasets, achieving improved survival prediction on TCGA-BLCA and competitive performance on TCGA-BRCA compared to state-of-the-art methods. This dual fusion strategy enhances interpretability and classification performance, highlighting its potential for clinical diagnostics.
Compositional Segmentation of Cardiac Images Leveraging Metadata
Oct 30, 2024Abstract:Cardiac image segmentation is essential for automated cardiac function assessment and monitoring of changes in cardiac structures over time. Inspired by coarse-to-fine approaches in image analysis, we propose a novel multitask compositional segmentation approach that can simultaneously localize the heart in a cardiac image and perform part-based segmentation of different regions of interest. We demonstrate that this compositional approach achieves better results than direct segmentation of the anatomies. Further, we propose a novel Cross-Modal Feature Integration (CMFI) module to leverage the metadata related to cardiac imaging collected during image acquisition. We perform experiments on two different modalities, MRI and ultrasound, using public datasets, Multi-disease, Multi-View, and Multi-Centre (M&Ms-2) and Multi-structure Ultrasound Segmentation (CAMUS) data, to showcase the efficiency of the proposed compositional segmentation method and Cross-Modal Feature Integration module incorporating metadata within the proposed compositional segmentation network. The source code is available: https://github.com/kabbas570/CompSeg-MetaData.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge