Abbas Khan
School of Electronic Engineering and Computer Science, Queen Mary University of London, UK, Queen Mary Digital Environment Research Institute
DVFL-Net: A Lightweight Distilled Video Focal Modulation Network for Spatio-Temporal Action Recognition
Jul 16, 2025Abstract:The landscape of video recognition has evolved significantly, shifting from traditional Convolutional Neural Networks (CNNs) to Transformer-based architectures for improved accuracy. While 3D CNNs have been effective at capturing spatiotemporal dynamics, recent Transformer models leverage self-attention to model long-range spatial and temporal dependencies. Despite achieving state-of-the-art performance on major benchmarks, Transformers remain computationally expensive, particularly with dense video data. To address this, we propose a lightweight Video Focal Modulation Network, DVFL-Net, which distills spatiotemporal knowledge from a large pre-trained teacher into a compact nano student model, enabling efficient on-device deployment. DVFL-Net utilizes knowledge distillation and spatial-temporal feature modulation to significantly reduce computation while preserving high recognition performance. We employ forward Kullback-Leibler (KL) divergence alongside spatio-temporal focal modulation to effectively transfer both local and global context from the Video-FocalNet Base (teacher) to the proposed VFL-Net (student). We evaluate DVFL-Net on UCF50, UCF101, HMDB51, SSV2, and Kinetics-400, benchmarking it against recent state-of-the-art methods in Human Action Recognition (HAR). Additionally, we conduct a detailed ablation study analyzing the impact of forward KL divergence. The results confirm the superiority of DVFL-Net in achieving an optimal balance between performance and efficiency, demonstrating lower memory usage, reduced GFLOPs, and strong accuracy, making it a practical solution for real-time HAR applications.
OD-VIRAT: A Large-Scale Benchmark for Object Detection in Realistic Surveillance Environments
Jul 16, 2025Abstract:Realistic human surveillance datasets are crucial for training and evaluating computer vision models under real-world conditions, facilitating the development of robust algorithms for human and human-interacting object detection in complex environments. These datasets need to offer diverse and challenging data to enable a comprehensive assessment of model performance and the creation of more reliable surveillance systems for public safety. To this end, we present two visual object detection benchmarks named OD-VIRAT Large and OD-VIRAT Tiny, aiming at advancing visual understanding tasks in surveillance imagery. The video sequences in both benchmarks cover 10 different scenes of human surveillance recorded from significant height and distance. The proposed benchmarks offer rich annotations of bounding boxes and categories, where OD-VIRAT Large has 8.7 million annotated instances in 599,996 images and OD-VIRAT Tiny has 288,901 annotated instances in 19,860 images. This work also focuses on benchmarking state-of-the-art object detection architectures, including RETMDET, YOLOX, RetinaNet, DETR, and Deformable-DETR on this object detection-specific variant of VIRAT dataset. To the best of our knowledge, it is the first work to examine the performance of these recently published state-of-the-art object detection architectures on realistic surveillance imagery under challenging conditions such as complex backgrounds, occluded objects, and small-scale objects. The proposed benchmarking and experimental settings will help in providing insights concerning the performance of selected object detection models and set the base for developing more efficient and robust object detection architectures.
Compositional Segmentation of Cardiac Images Leveraging Metadata
Oct 30, 2024Abstract:Cardiac image segmentation is essential for automated cardiac function assessment and monitoring of changes in cardiac structures over time. Inspired by coarse-to-fine approaches in image analysis, we propose a novel multitask compositional segmentation approach that can simultaneously localize the heart in a cardiac image and perform part-based segmentation of different regions of interest. We demonstrate that this compositional approach achieves better results than direct segmentation of the anatomies. Further, we propose a novel Cross-Modal Feature Integration (CMFI) module to leverage the metadata related to cardiac imaging collected during image acquisition. We perform experiments on two different modalities, MRI and ultrasound, using public datasets, Multi-disease, Multi-View, and Multi-Centre (M&Ms-2) and Multi-structure Ultrasound Segmentation (CAMUS) data, to showcase the efficiency of the proposed compositional segmentation method and Cross-Modal Feature Integration module incorporating metadata within the proposed compositional segmentation network. The source code is available: https://github.com/kabbas570/CompSeg-MetaData.
Convolution and Attention-Free Mamba-based Cardiac Image Segmentation
Jun 09, 2024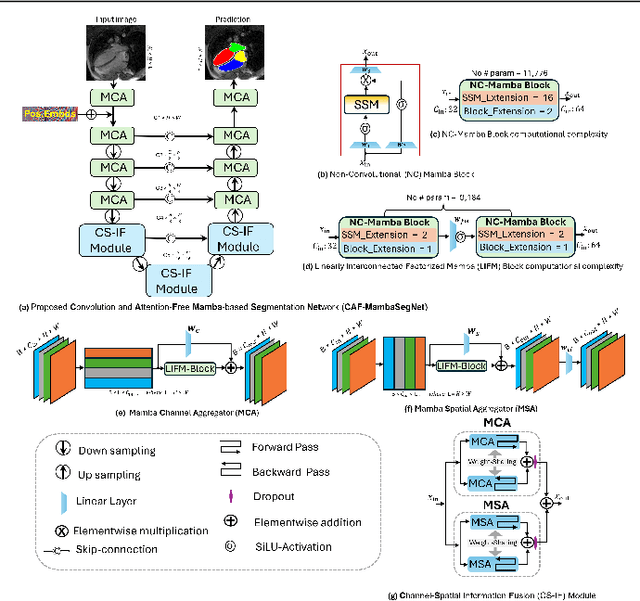
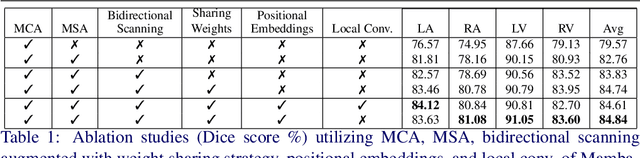
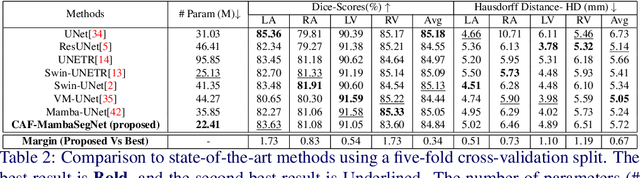
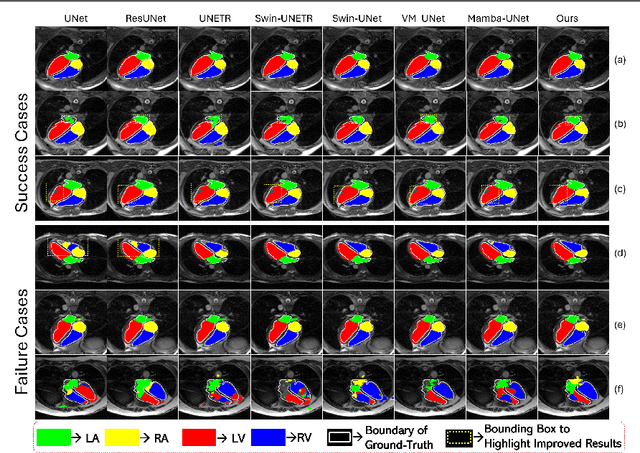
Abstract:Convolutional Neural Networks (CNNs) and Transformer-based self-attention models have become standard for medical image segmentation. This paper demonstrates that convolution and self-attention, while widely used, are not the only effective methods for segmentation. Breaking with convention, we present a Convolution and self-Attention Free Mamba-based semantic Segmentation Network named CAF-MambaSegNet. Specifically, we design a Mamba-based Channel Aggregator and Spatial Aggregator, which are applied independently in each encoder-decoder stage. The Channel Aggregator extracts information across different channels, and the Spatial Aggregator learns features across different spatial locations. We also propose a Linearly Interconnected Factorized Mamba (LIFM) Block to reduce the computational complexity of a Mamba and to enhance its decision function by introducing a non-linearity between two factorized Mamba blocks. Our goal is not to outperform state-of-the-art results but to show how this innovative, convolution and self-attention-free method can inspire further research beyond well-established CNNs and Transformers, achieving linear complexity and reducing the number of parameters. Source code and pre-trained models will be publicly available.
Multi-view Cardiac Image Segmentation via Trans-Dimensional Priors
Apr 25, 2024



Abstract:We propose a novel multi-stage trans-dimensional architecture for multi-view cardiac image segmentation. Our method exploits the relationship between long-axis (2D) and short-axis (3D) magnetic resonance (MR) images to perform a sequential 3D-to-2D-to-3D segmentation, segmenting the long-axis and short-axis images. In the first stage, 3D segmentation is performed using the short-axis image, and the prediction is transformed to the long-axis view and used as a segmentation prior in the next stage. In the second step, the heart region is localized and cropped around the segmentation prior using a Heart Localization and Cropping (HLC) module, focusing the subsequent model on the heart region of the image, where a 2D segmentation is performed. Similarly, we transform the long-axis prediction to the short-axis view, localize and crop the heart region and again perform a 3D segmentation to refine the initial short-axis segmentation. We evaluate our proposed method on the Multi-Disease, Multi-View & Multi-Center Right Ventricular Segmentation in Cardiac MRI (M&Ms-2) dataset, where our method outperforms state-of-the-art methods in segmenting cardiac regions of interest in both short-axis and long-axis images. The pre-trained models, source code, and implementation details will be publicly available.
MOAB: Multi-Modal Outer Arithmetic Block For Fusion Of Histopathological Images And Genetic Data For Brain Tumor Grading
Mar 11, 2024Abstract:Brain tumors are an abnormal growth of cells in the brain. They can be classified into distinct grades based on their growth. Often grading is performed based on a histological image and is one of the most significant predictors of a patients prognosis, the higher the grade, the more aggressive the tumor. Correct diagnosis of a tumor grade remains challenging. Though histopathological grading has been shown to be prognostic, results are subject to interobserver variability, even among experienced pathologists. Recently, the World Health Organization reported that advances in molecular genetics have led to improvements in tumor classification. This paper seeks to integrate histological images and genetic data for improved computer-aided diagnosis. We propose a novel Multi-modal Outer Arithmetic Block (MOAB) based on arithmetic operations to combine latent representations of the different modalities for predicting the tumor grade (Grade \rom{2}, \rom{3} and \rom{4}). Extensive experiments evaluate the effectiveness of our approach. By applying MOAB to The Cancer Genome Atlas (TCGA) glioma dataset, we show that it can improve separation between similar classes (Grade \rom{2} and \rom{3}) and outperform prior state-of-the-art grade classification techniques.
Crop and Couple: cardiac image segmentation using interlinked specialist networks
Feb 14, 2024Abstract:Diagnosis of cardiovascular disease using automated methods often relies on the critical task of cardiac image segmentation. We propose a novel strategy that performs segmentation using specialist networks that focus on a single anatomy (left ventricle, right ventricle, or myocardium). Given an input long-axis cardiac MR image, our method performs a ternary segmentation in the first stage to identify these anatomical regions, followed by cropping the original image to focus subsequent processing on the anatomical regions. The specialist networks are coupled through an attention mechanism that performs cross-attention to interlink features from different anatomies, serving as a soft relative shape prior. Central to our approach is an additive attention block (E-2A block), which is used throughout our architecture thanks to its efficiency.
Automated segmentation of rheumatoid arthritis immunohistochemistry stained synovial tissue
Sep 13, 2023Abstract:Rheumatoid Arthritis (RA) is a chronic, autoimmune disease which primarily affects the joint's synovial tissue. It is a highly heterogeneous disease, with wide cellular and molecular variability observed in synovial tissues. Over the last two decades, the methods available for their study have advanced considerably. In particular, Immunohistochemistry stains are well suited to highlighting the functional organisation of samples. Yet, analysis of IHC-stained synovial tissue samples is still overwhelmingly done manually and semi-quantitatively by expert pathologists. This is because in addition to the fragmented nature of IHC stained synovial tissue, there exist wide variations in intensity and colour, strong clinical centre batch effect, as well as the presence of many undesirable artefacts present in gigapixel Whole Slide Images (WSIs), such as water droplets, pen annotation, folded tissue, blurriness, etc. There is therefore a strong need for a robust, repeatable automated tissue segmentation algorithm which can cope with this variability and provide support to imaging pipelines. We train a UNET on a hand-curated, heterogeneous real-world multi-centre clinical dataset R4RA, which contains multiple types of IHC staining. The model obtains a DICE score of 0.865 and successfully segments different types of IHC staining, as well as dealing with variance in colours, intensity and common WSIs artefacts from the different clinical centres. It can be used as the first step in an automated image analysis pipeline for synovial tissue samples stained with IHC, increasing speed, reproducibility and robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge