Martin Benning
Multi-Frame Blind Manifold Deconvolution for Rotating Synthetic Aperture Imaging
Jan 31, 2025Abstract:Rotating synthetic aperture (RSA) imaging system captures images of the target scene at different rotation angles by rotating a rectangular aperture. Deblurring acquired RSA images plays a critical role in reconstructing a latent sharp image underlying the scene. In the past decade, the emergence of blind convolution technology has revolutionised this field by its ability to model complex features from acquired images. Most of the existing methods attempt to solve the above ill-posed inverse problem through maximising a posterior. Despite this progress, researchers have paid limited attention to exploring low-dimensional manifold structures of the latent image within a high-dimensional ambient-space. Here, we propose a novel method to process RSA images using manifold fitting and penalisation in the content of multi-frame blind convolution. We develop fast algorithms for implementing the proposed procedure. Simulation studies demonstrate that manifold-based deconvolution can outperform conventional deconvolution algorithms in the sense that it can generate a sharper estimate of the latent image in terms of estimating pixel intensities and preserving structural details.
Compositional Segmentation of Cardiac Images Leveraging Metadata
Oct 30, 2024Abstract:Cardiac image segmentation is essential for automated cardiac function assessment and monitoring of changes in cardiac structures over time. Inspired by coarse-to-fine approaches in image analysis, we propose a novel multitask compositional segmentation approach that can simultaneously localize the heart in a cardiac image and perform part-based segmentation of different regions of interest. We demonstrate that this compositional approach achieves better results than direct segmentation of the anatomies. Further, we propose a novel Cross-Modal Feature Integration (CMFI) module to leverage the metadata related to cardiac imaging collected during image acquisition. We perform experiments on two different modalities, MRI and ultrasound, using public datasets, Multi-disease, Multi-View, and Multi-Centre (M&Ms-2) and Multi-structure Ultrasound Segmentation (CAMUS) data, to showcase the efficiency of the proposed compositional segmentation method and Cross-Modal Feature Integration module incorporating metadata within the proposed compositional segmentation network. The source code is available: https://github.com/kabbas570/CompSeg-MetaData.
A lifted Bregman strategy for training unfolded proximal neural network Gaussian denoisers
Aug 16, 2024Abstract:Unfolded proximal neural networks (PNNs) form a family of methods that combines deep learning and proximal optimization approaches. They consist in designing a neural network for a specific task by unrolling a proximal algorithm for a fixed number of iterations, where linearities can be learned from prior training procedure. PNNs have shown to be more robust than traditional deep learning approaches while reaching at least as good performances, in particular in computational imaging. However, training PNNs still depends on the efficiency of available training algorithms. In this work, we propose a lifted training formulation based on Bregman distances for unfolded PNNs. Leveraging the deterministic mini-batch block-coordinate forward-backward method, we design a bespoke computational strategy beyond traditional back-propagation methods for solving the resulting learning problem efficiently. We assess the behaviour of the proposed training approach for PNNs through numerical simulations on image denoising, considering a denoising PNN whose structure is based on dual proximal-gradient iterations.
Improving Interpretability and Robustness for the Detection of AI-Generated Images
Jun 21, 2024



Abstract:With growing abilities of generative models, artificial content detection becomes an increasingly important and difficult task. However, all popular approaches to this problem suffer from poor generalization across domains and generative models. In this work, we focus on the robustness of AI-generated image (AIGI) detectors. We analyze existing state-of-the-art AIGI detection methods based on frozen CLIP embeddings and show how to interpret them, shedding light on how images produced by various AI generators differ from real ones. Next we propose two ways to improve robustness: based on removing harmful components of the embedding vector and based on selecting the best performing attention heads in the image encoder model. Our methods increase the mean out-of-distribution (OOD) classification score by up to 6% for cross-model transfer. We also propose a new dataset for AIGI detection and use it in our evaluation; we believe this dataset will help boost further research. The dataset and code are provided as a supplement.
Convolution and Attention-Free Mamba-based Cardiac Image Segmentation
Jun 09, 2024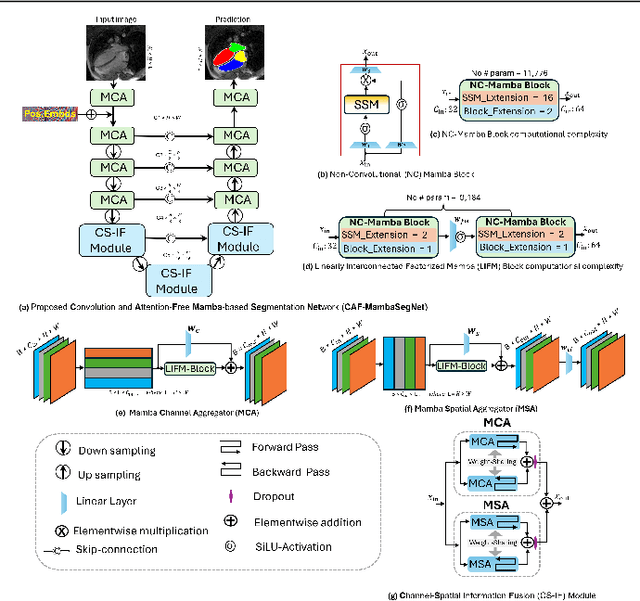
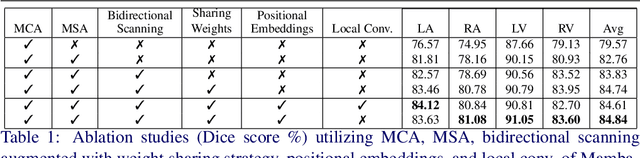
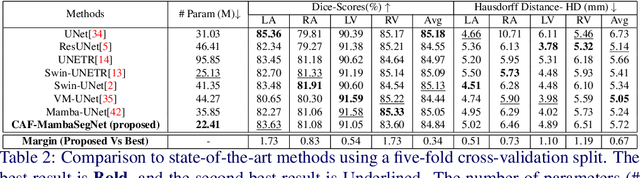
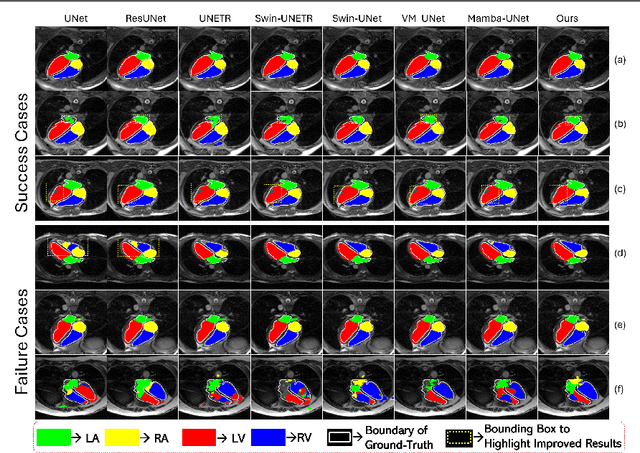
Abstract:Convolutional Neural Networks (CNNs) and Transformer-based self-attention models have become standard for medical image segmentation. This paper demonstrates that convolution and self-attention, while widely used, are not the only effective methods for segmentation. Breaking with convention, we present a Convolution and self-Attention Free Mamba-based semantic Segmentation Network named CAF-MambaSegNet. Specifically, we design a Mamba-based Channel Aggregator and Spatial Aggregator, which are applied independently in each encoder-decoder stage. The Channel Aggregator extracts information across different channels, and the Spatial Aggregator learns features across different spatial locations. We also propose a Linearly Interconnected Factorized Mamba (LIFM) Block to reduce the computational complexity of a Mamba and to enhance its decision function by introducing a non-linearity between two factorized Mamba blocks. Our goal is not to outperform state-of-the-art results but to show how this innovative, convolution and self-attention-free method can inspire further research beyond well-established CNNs and Transformers, achieving linear complexity and reducing the number of parameters. Source code and pre-trained models will be publicly available.
Multi-view Cardiac Image Segmentation via Trans-Dimensional Priors
Apr 25, 2024



Abstract:We propose a novel multi-stage trans-dimensional architecture for multi-view cardiac image segmentation. Our method exploits the relationship between long-axis (2D) and short-axis (3D) magnetic resonance (MR) images to perform a sequential 3D-to-2D-to-3D segmentation, segmenting the long-axis and short-axis images. In the first stage, 3D segmentation is performed using the short-axis image, and the prediction is transformed to the long-axis view and used as a segmentation prior in the next stage. In the second step, the heart region is localized and cropped around the segmentation prior using a Heart Localization and Cropping (HLC) module, focusing the subsequent model on the heart region of the image, where a 2D segmentation is performed. Similarly, we transform the long-axis prediction to the short-axis view, localize and crop the heart region and again perform a 3D segmentation to refine the initial short-axis segmentation. We evaluate our proposed method on the Multi-Disease, Multi-View & Multi-Center Right Ventricular Segmentation in Cardiac MRI (M&Ms-2) dataset, where our method outperforms state-of-the-art methods in segmenting cardiac regions of interest in both short-axis and long-axis images. The pre-trained models, source code, and implementation details will be publicly available.
RAVE: Residual Vector Embedding for CLIP-Guided Backlit Image Enhancement
Apr 03, 2024Abstract:In this paper we propose a novel modification of Contrastive Language-Image Pre-Training (CLIP) guidance for the task of unsupervised backlit image enhancement. Our work builds on the state-of-the-art CLIP-LIT approach, which learns a prompt pair by constraining the text-image similarity between a prompt (negative/positive sample) and a corresponding image (backlit image/well-lit image) in the CLIP embedding space. Learned prompts then guide an image enhancement network. Based on the CLIP-LIT framework, we propose two novel methods for CLIP guidance. First, we show that instead of tuning prompts in the space of text embeddings, it is possible to directly tune their embeddings in the latent space without any loss in quality. This accelerates training and potentially enables the use of additional encoders that do not have a text encoder. Second, we propose a novel approach that does not require any prompt tuning. Instead, based on CLIP embeddings of backlit and well-lit images from training data, we compute the residual vector in the embedding space as a simple difference between the mean embeddings of the well-lit and backlit images. This vector then guides the enhancement network during training, pushing a backlit image towards the space of well-lit images. This approach further dramatically reduces training time, stabilizes training and produces high quality enhanced images without artifacts, both in supervised and unsupervised training regimes. Additionally, we show that residual vectors can be interpreted, revealing biases in training data, and thereby enabling potential bias correction.
Crop and Couple: cardiac image segmentation using interlinked specialist networks
Feb 14, 2024Abstract:Diagnosis of cardiovascular disease using automated methods often relies on the critical task of cardiac image segmentation. We propose a novel strategy that performs segmentation using specialist networks that focus on a single anatomy (left ventricle, right ventricle, or myocardium). Given an input long-axis cardiac MR image, our method performs a ternary segmentation in the first stage to identify these anatomical regions, followed by cropping the original image to focus subsequent processing on the anatomical regions. The specialist networks are coupled through an attention mechanism that performs cross-attention to interlink features from different anatomies, serving as a soft relative shape prior. Central to our approach is an additive attention block (E-2A block), which is used throughout our architecture thanks to its efficiency.
A Lifted Bregman Formulation for the Inversion of Deep Neural Networks
Mar 01, 2023Abstract:We propose a novel framework for the regularised inversion of deep neural networks. The framework is based on the authors' recent work on training feed-forward neural networks without the differentiation of activation functions. The framework lifts the parameter space into a higher dimensional space by introducing auxiliary variables, and penalises these variables with tailored Bregman distances. We propose a family of variational regularisations based on these Bregman distances, present theoretical results and support their practical application with numerical examples. In particular, we present the first convergence result (to the best of our knowledge) for the regularised inversion of a single-layer perceptron that only assumes that the solution of the inverse problem is in the range of the regularisation operator, and that shows that the regularised inverse provably converges to the true inverse if measurement errors converge to zero.
Convergent Data-driven Regularizations for CT Reconstruction
Dec 14, 2022Abstract:The reconstruction of images from their corresponding noisy Radon transform is a typical example of an ill-posed linear inverse problem as arising in the application of computerized tomography (CT). As the (na\"{\i}ve) solution does not depend on the measured data continuously, regularization is needed to re-establish a continuous dependence. In this work, we investigate simple, but yet still provably convergent approaches to learning linear regularization methods from data. More specifically, we analyze two approaches: One generic linear regularization that learns how to manipulate the singular values of the linear operator in an extension of [1], and one tailored approach in the Fourier domain that is specific to CT-reconstruction. We prove that such approaches become convergent regularization methods as well as the fact that the reconstructions they provide are typically much smoother than the training data they were trained on. Finally, we compare the spectral as well as the Fourier-based approaches for CT-reconstruction numerically, discuss their advantages and disadvantages and investigate the effect of discretization errors at different resolutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge