Omnia Alwazzan
School of Electronic Engineering and Computer Science, Queen Mary University of London, UK, Queen Mary Digital Environment Research Institute
Towards deployment-centric multimodal AI beyond vision and language
Apr 04, 2025Abstract:Multimodal artificial intelligence (AI) integrates diverse types of data via machine learning to improve understanding, prediction, and decision-making across disciplines such as healthcare, science, and engineering. However, most multimodal AI advances focus on models for vision and language data, while their deployability remains a key challenge. We advocate a deployment-centric workflow that incorporates deployment constraints early to reduce the likelihood of undeployable solutions, complementing data-centric and model-centric approaches. We also emphasise deeper integration across multiple levels of multimodality and multidisciplinary collaboration to significantly broaden the research scope beyond vision and language. To facilitate this approach, we identify common multimodal-AI-specific challenges shared across disciplines and examine three real-world use cases: pandemic response, self-driving car design, and climate change adaptation, drawing expertise from healthcare, social science, engineering, science, sustainability, and finance. By fostering multidisciplinary dialogue and open research practices, our community can accelerate deployment-centric development for broad societal impact.
BioX-CPath: Biologically-driven Explainable Diagnostics for Multistain IHC Computational Pathology
Mar 26, 2025



Abstract:The development of biologically interpretable and explainable models remains a key challenge in computational pathology, particularly for multistain immunohistochemistry (IHC) analysis. We present BioX-CPath, an explainable graph neural network architecture for whole slide image (WSI) classification that leverages both spatial and semantic features across multiple stains. At its core, BioX-CPath introduces a novel Stain-Aware Attention Pooling (SAAP) module that generates biologically meaningful, stain-aware patient embeddings. Our approach achieves state-of-the-art performance on both Rheumatoid Arthritis and Sjogren's Disease multistain datasets. Beyond performance metrics, BioX-CPath provides interpretable insights through stain attention scores, entropy measures, and stain interaction scores, that permit measuring model alignment with known pathological mechanisms. This biological grounding, combined with strong classification performance, makes BioX-CPath particularly suitable for clinical applications where interpretability is key. Source code and documentation can be found at: https://github.com/AmayaGS/BioX-CPath.
Multimodal Outer Arithmetic Block Dual Fusion of Whole Slide Images and Omics Data for Precision Oncology
Nov 26, 2024Abstract:Developing a central nervous system (CNS) tumor classifier by integrating DNA methylation data with Whole Slide Images (WSI) offers significant potential for enhancing diagnostic precision in neuropathology. Existing approaches typically integrate encoded omic data with histology only once - either at an early or late fusion stage - while reintroducing encoded omic data to create a dual fusion variant remains unexplored. Nevertheless, reintroduction of omic embeddings during early and late fusion enables the capture of complementary information from localized patch-level and holistic slide-level interactions, allowing boosted performance through advanced multimodal integration. To achieve this, we propose a dual fusion framework that integrates omic data at both early and late stages, fully leveraging its diagnostic strength. In the early fusion stage, omic embeddings are projected into a patch-wise latent space, generating omic-WSI embeddings that encapsulate per-patch molecular and morphological insights, effectively incorporating this information into the spatial representation of histology. These embeddings are refined with a multiple instance learning gated attention mechanism to attend to critical patches. In the late fusion stage, we reintroduce the omic data by fusing it with slide-level omic-WSI embeddings using a Multimodal Outer Arithmetic Block (MOAB), which richly intermingles features from both modalities, capturing their global correlations and complementarity. We demonstrate accurate CNS tumor subtyping across 20 fine-grained subtypes and validate our approach on benchmark datasets, achieving improved survival prediction on TCGA-BLCA and competitive performance on TCGA-BRCA compared to state-of-the-art methods. This dual fusion strategy enhances interpretability and classification performance, highlighting its potential for clinical diagnostics.
MOAB: Multi-Modal Outer Arithmetic Block For Fusion Of Histopathological Images And Genetic Data For Brain Tumor Grading
Mar 11, 2024Abstract:Brain tumors are an abnormal growth of cells in the brain. They can be classified into distinct grades based on their growth. Often grading is performed based on a histological image and is one of the most significant predictors of a patients prognosis, the higher the grade, the more aggressive the tumor. Correct diagnosis of a tumor grade remains challenging. Though histopathological grading has been shown to be prognostic, results are subject to interobserver variability, even among experienced pathologists. Recently, the World Health Organization reported that advances in molecular genetics have led to improvements in tumor classification. This paper seeks to integrate histological images and genetic data for improved computer-aided diagnosis. We propose a novel Multi-modal Outer Arithmetic Block (MOAB) based on arithmetic operations to combine latent representations of the different modalities for predicting the tumor grade (Grade \rom{2}, \rom{3} and \rom{4}). Extensive experiments evaluate the effectiveness of our approach. By applying MOAB to The Cancer Genome Atlas (TCGA) glioma dataset, we show that it can improve separation between similar classes (Grade \rom{2} and \rom{3}) and outperform prior state-of-the-art grade classification techniques.
FOAA: Flattened Outer Arithmetic Attention For Multimodal Tumor Classification
Mar 10, 2024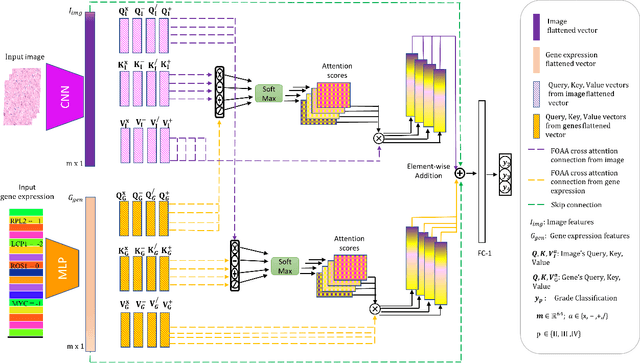
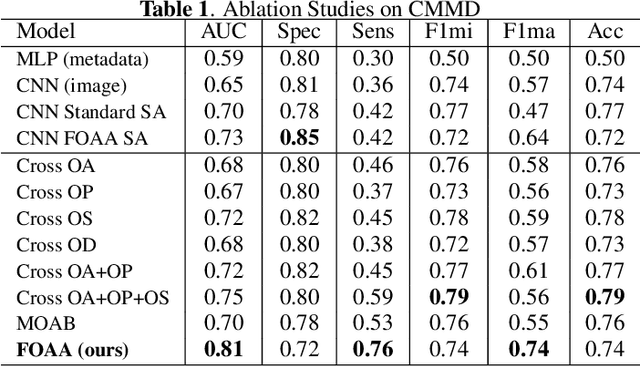
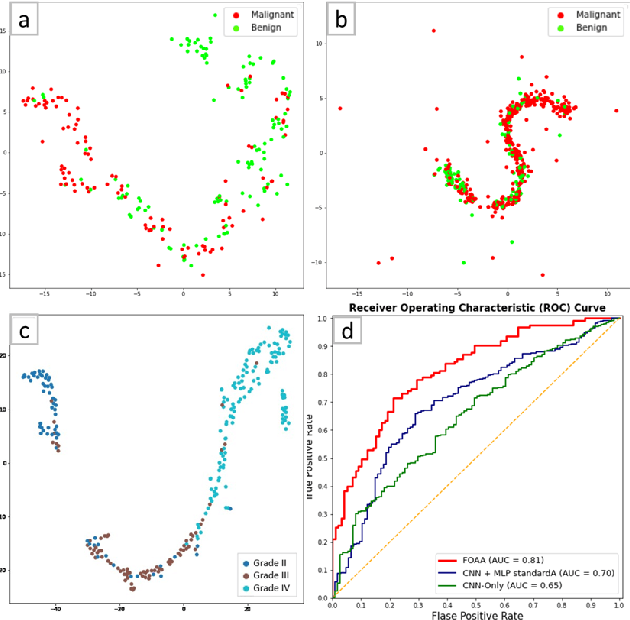
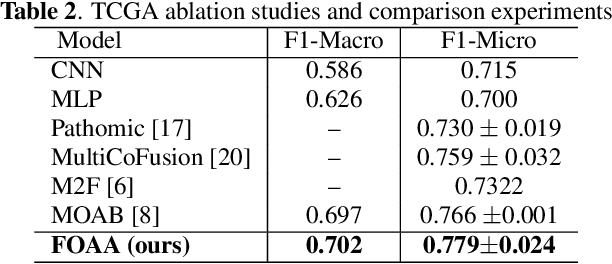
Abstract:Fusion of multimodal healthcare data holds great promise to provide a holistic view of a patient's health, taking advantage of the complementarity of different modalities while leveraging their correlation. This paper proposes a simple and effective approach, inspired by attention, to fuse discriminative features from different modalities. We propose a novel attention mechanism, called Flattened Outer Arithmetic Attention (FOAA), which relies on outer arithmetic operators (addition, subtraction, product, and division) to compute attention scores from keys, queries and values derived from flattened embeddings of each modality. We demonstrate how FOAA can be implemented for self-attention and cross-attention, providing a reusable component in neural network architectures. We evaluate FOAA on two datasets for multimodal tumor classification and achieve state-of-the-art results, and we demonstrate that features enriched by FOAA are superior to those derived from other fusion approaches. The code is publicly available at \href{https://github.com/omniaalwazzan/FOAA}{https://github.com/omniaalwazzan/FOAA}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge