Yuqi Fang
Controlled Self-Evolution for Algorithmic Code Optimization
Jan 13, 2026Abstract:Self-evolution methods enhance code generation through iterative "generate-verify-refine" cycles, yet existing approaches suffer from low exploration efficiency, failing to discover solutions with superior complexity within limited budgets. This inefficiency stems from initialization bias trapping evolution in poor solution regions, uncontrolled stochastic operations lacking feedback guidance, and insufficient experience utilization across tasks. To address these bottlenecks, we propose Controlled Self-Evolution (CSE), which consists of three key components. Diversified Planning Initialization generates structurally distinct algorithmic strategies for broad solution space coverage. Genetic Evolution replaces stochastic operations with feedback-guided mechanisms, enabling targeted mutation and compositional crossover. Hierarchical Evolution Memory captures both successful and failed experiences at inter-task and intra-task levels. Experiments on EffiBench-X demonstrate that CSE consistently outperforms all baselines across various LLM backbones. Furthermore, CSE achieves higher efficiency from early generations and maintains continuous improvement throughout evolution. Our code is publicly available at https://github.com/QuantaAlpha/EvoControl.
ERSR: An Ellipse-constrained pseudo-label refinement and symmetric regularization framework for semi-supervised fetal head segmentation in ultrasound images
Aug 27, 2025
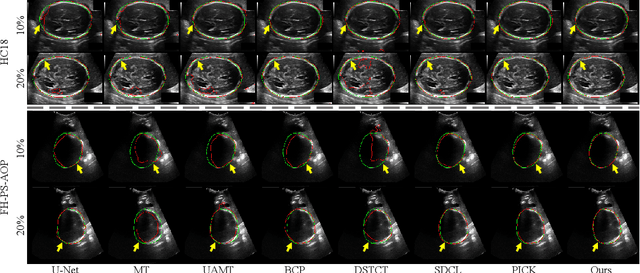

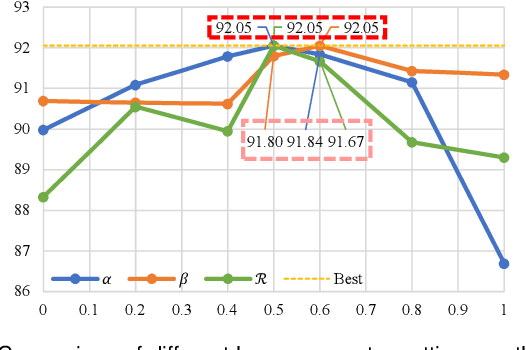
Abstract:Automated segmentation of the fetal head in ultrasound images is critical for prenatal monitoring. However, achieving robust segmentation remains challenging due to the poor quality of ultrasound images and the lack of annotated data. Semi-supervised methods alleviate the lack of annotated data but struggle with the unique characteristics of fetal head ultrasound images, making it challenging to generate reliable pseudo-labels and enforce effective consistency regularization constraints. To address this issue, we propose a novel semi-supervised framework, ERSR, for fetal head ultrasound segmentation. Our framework consists of the dual-scoring adaptive filtering strategy, the ellipse-constrained pseudo-label refinement, and the symmetry-based multiple consistency regularization. The dual-scoring adaptive filtering strategy uses boundary consistency and contour regularity criteria to evaluate and filter teacher outputs. The ellipse-constrained pseudo-label refinement refines these filtered outputs by fitting least-squares ellipses, which strengthens pixels near the center of the fitted ellipse and suppresses noise simultaneously. The symmetry-based multiple consistency regularization enforces multi-level consistency across perturbed images, symmetric regions, and between original predictions and pseudo-labels, enabling the model to capture robust and stable shape representations. Our method achieves state-of-the-art performance on two benchmarks. On the HC18 dataset, it reaches Dice scores of 92.05% and 95.36% with 10% and 20% labeled data, respectively. On the PSFH dataset, the scores are 91.68% and 93.70% under the same settings.
Topology-Aware Graph Augmentation for Predicting Clinical Trajectories in Neurocognitive Disorders
Oct 31, 2024



Abstract:Brain networks/graphs derived from resting-state functional MRI (fMRI) help study underlying pathophysiology of neurocognitive disorders by measuring neuronal activities in the brain. Some studies utilize learning-based methods for brain network analysis, but typically suffer from low model generalizability caused by scarce labeled fMRI data. As a notable self-supervised strategy, graph contrastive learning helps leverage auxiliary unlabeled data. But existing methods generally arbitrarily perturb graph nodes/edges to generate augmented graphs, without considering essential topology information of brain networks. To this end, we propose a topology-aware graph augmentation (TGA) framework, comprising a pretext model to train a generalizable encoder on large-scale unlabeled fMRI cohorts and a task-specific model to perform downstream tasks on a small target dataset. In the pretext model, we design two novel topology-aware graph augmentation strategies: (1) hub-preserving node dropping that prioritizes preserving brain hub regions according to node importance, and (2) weight-dependent edge removing that focuses on keeping important functional connectivities based on edge weights. Experiments on 1, 688 fMRI scans suggest that TGA outperforms several state-of-the-art methods.
ACTION: Augmentation and Computation Toolbox for Brain Network Analysis with Functional MRI
May 10, 2024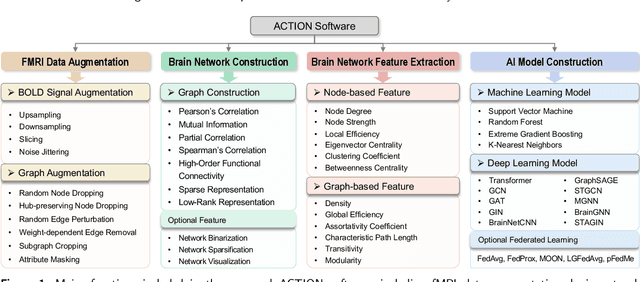
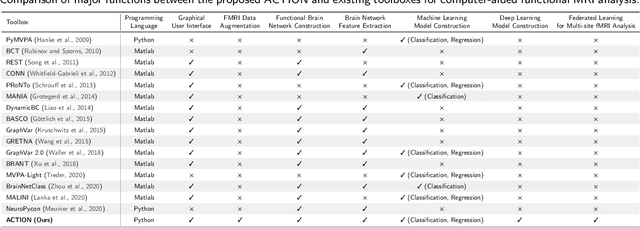
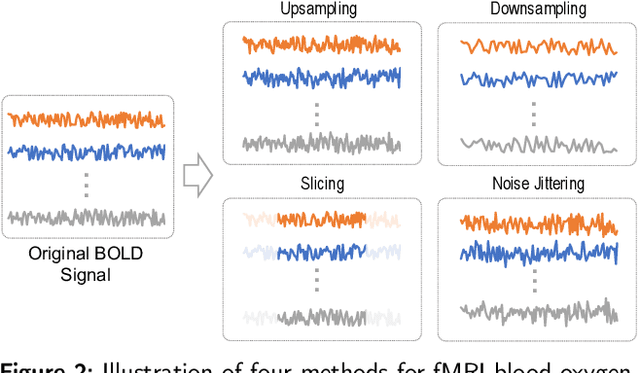
Abstract:Functional magnetic resonance imaging (fMRI) has been increasingly employed to investigate functional brain activity. Many fMRI-related software/toolboxes have been developed, providing specialized algorithms for fMRI analysis. However, existing toolboxes seldom consider fMRI data augmentation, which is quite useful, especially in studies with limited or imbalanced data. Moreover, current studies usually focus on analyzing fMRI using conventional machine learning models that rely on human-engineered fMRI features, without investigating deep learning models that can automatically learn data-driven fMRI representations. In this work, we develop an open-source toolbox, called Augmentation and Computation Toolbox for braIn netwOrk aNalysis (ACTION), offering comprehensive functions to streamline fMRI analysis. The ACTION is a Python-based and cross-platform toolbox with graphical user-friendly interfaces. It enables automatic fMRI augmentation, covering blood-oxygen-level-dependent (BOLD) signal augmentation and brain network augmentation. Many popular methods for brain network construction and network feature extraction are included. In particular, it supports constructing deep learning models, which leverage large-scale auxiliary unlabeled data (3,800+ resting-state fMRI scans) for model pretraining to enhance model performance for downstream tasks. To facilitate multi-site fMRI studies, it is also equipped with several popular federated learning strategies. Furthermore, it enables users to design and test custom algorithms through scripting, greatly improving its utility and extensibility. We demonstrate the effectiveness and user-friendliness of ACTION on real fMRI data and present the experimental results. The software, along with its source code and manual, can be accessed online.
Source-Free Collaborative Domain Adaptation via Multi-Perspective Feature Enrichment for Functional MRI Analysis
Aug 24, 2023



Abstract:Resting-state functional MRI (rs-fMRI) is increasingly employed in multi-site research to aid neurological disorder analysis. Existing studies usually suffer from significant cross-site/domain data heterogeneity caused by site effects such as differences in scanners/protocols. Many methods have been proposed to reduce fMRI heterogeneity between source and target domains, heavily relying on the availability of source data. But acquiring source data is challenging due to privacy concerns and/or data storage burdens in multi-site studies. To this end, we design a source-free collaborative domain adaptation (SCDA) framework for fMRI analysis, where only a pretrained source model and unlabeled target data are accessible. Specifically, a multi-perspective feature enrichment method (MFE) is developed for target fMRI analysis, consisting of multiple collaborative branches to dynamically capture fMRI features of unlabeled target data from multiple views. Each branch has a data-feeding module, a spatiotemporal feature encoder, and a class predictor. A mutual-consistency constraint is designed to encourage pair-wise consistency of latent features of the same input generated from these branches for robust representation learning. To facilitate efficient cross-domain knowledge transfer without source data, we initialize MFE using parameters of a pretrained source model. We also introduce an unsupervised pretraining strategy using 3,806 unlabeled fMRIs from three large-scale auxiliary databases, aiming to obtain a general feature encoder. Experimental results on three public datasets and one private dataset demonstrate the efficacy of our method in cross-scanner and cross-study prediction tasks. The model pretrained on large-scale rs-fMRI data has been released to the public.
Leveraging Brain Modularity Prior for Interpretable Representation Learning of fMRI
Jun 24, 2023Abstract:Resting-state functional magnetic resonance imaging (rs-fMRI) can reflect spontaneous neural activities in brain and is widely used for brain disorder analysis.Previous studies propose to extract fMRI representations through diverse machine/deep learning methods for subsequent analysis. But the learned features typically lack biological interpretability, which limits their clinical utility. From the view of graph theory, the brain exhibits a remarkable modular structure in spontaneous brain functional networks, with each module comprised of functionally interconnected brain regions-of-interest (ROIs). However, most existing learning-based methods for fMRI analysis fail to adequately utilize such brain modularity prior. In this paper, we propose a Brain Modularity-constrained dynamic Representation learning (BMR) framework for interpretable fMRI analysis, consisting of three major components: (1) dynamic graph construction, (2) dynamic graph learning via a novel modularity-constrained graph neural network(MGNN), (3) prediction and biomarker detection for interpretable fMRI analysis. Especially, three core neurocognitive modules (i.e., salience network, central executive network, and default mode network) are explicitly incorporated into the MGNN, encouraging the nodes/ROIs within the same module to share similar representations. To further enhance discriminative ability of learned features, we also encourage the MGNN to preserve the network topology of input graphs via a graph topology reconstruction constraint. Experimental results on 534 subjects with rs-fMRI scans from two datasets validate the effectiveness of the proposed method. The identified discriminative brain ROIs and functional connectivities can be regarded as potential fMRI biomarkers to aid in clinical diagnosis.
Source-Free Unsupervised Domain Adaptation: A Survey
Jan 06, 2023



Abstract:Unsupervised domain adaptation (UDA) via deep learning has attracted appealing attention for tackling domain-shift problems caused by distribution discrepancy across different domains. Existing UDA approaches highly depend on the accessibility of source domain data, which is usually limited in practical scenarios due to privacy protection, data storage and transmission cost, and computation burden. To tackle this issue, many source-free unsupervised domain adaptation (SFUDA) methods have been proposed recently, which perform knowledge transfer from a pre-trained source model to unlabeled target domain with source data inaccessible. A comprehensive review of these works on SFUDA is of great significance. In this paper, we provide a timely and systematic literature review of existing SFUDA approaches from a technical perspective. Specifically, we categorize current SFUDA studies into two groups, i.e., white-box SFUDA and black-box SFUDA, and further divide them into finer subcategories based on different learning strategies they use. We also investigate the challenges of methods in each subcategory, discuss the advantages/disadvantages of white-box and black-box SFUDA methods, conclude the commonly used benchmark datasets, and summarize the popular techniques for improved generalizability of models learned without using source data. We finally discuss several promising future directions in this field.
DeepNoise: Disentanglement of Experimental Noise from Real Biological Signals based on Fluorescent Microscopy Image Classification via Deep Learning
Sep 13, 2022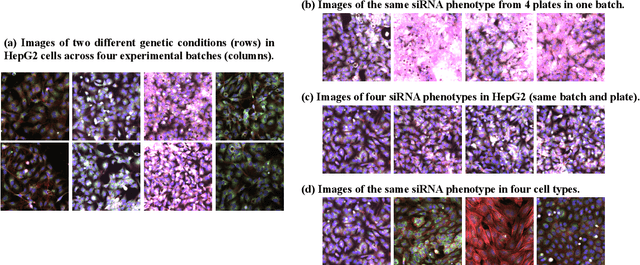
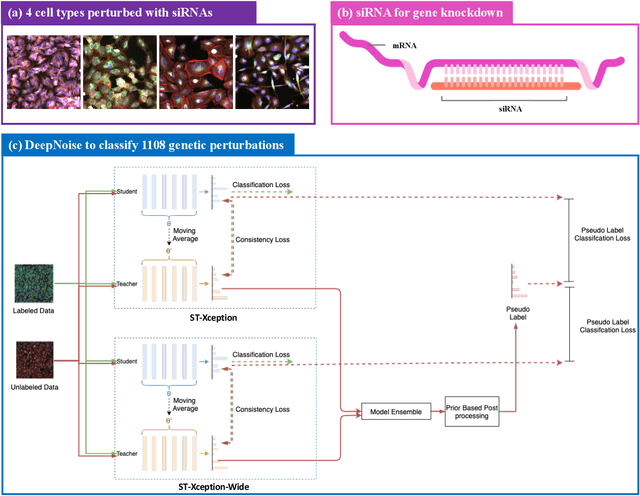
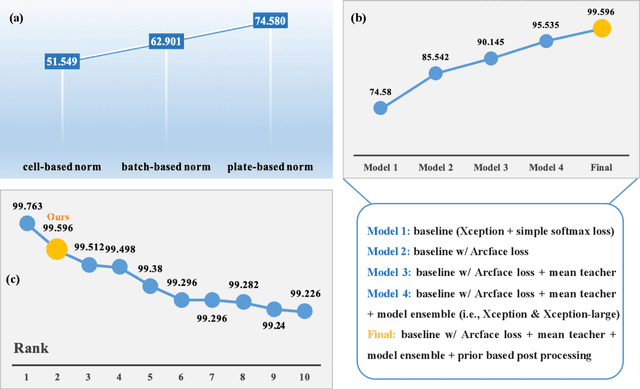
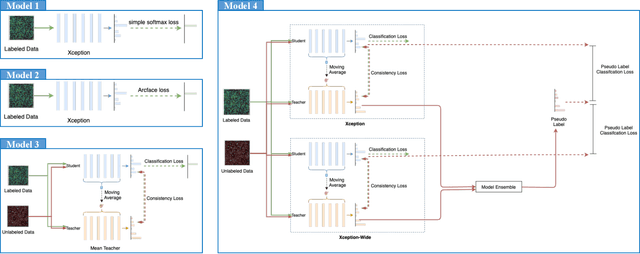
Abstract:The high-content image-based assay is commonly leveraged for identifying the phenotypic impact of genetic perturbations in biology field. However, a persistent issue remains unsolved during experiments: the interferential technical noise caused by systematic errors (e.g., temperature, reagent concentration, and well location) is always mixed up with the real biological signals, leading to misinterpretation of any conclusion drawn. Here, we show a mean teacher based deep learning model (DeepNoise) that can disentangle biological signals from the experimental noise. Specifically, we aim to classify the phenotypic impact of 1,108 different genetic perturbations screened from 125,510 fluorescent microscopy images, which are totally unrecognizable by human eye. We validate our model by participating in the Recursion Cellular Image Classification Challenge, and our proposed method achieves an extremely high classification score (Acc: 99.596%), ranking the 2nd place among 866 participating groups. This promising result indicates the successful separation of biological and technical factors, which might help decrease the cost of treatment development and expedite the drug discovery process.
A Survey on Deep-Learning Approaches for Vehicle Trajectory Prediction in Autonomous Driving
Oct 29, 2021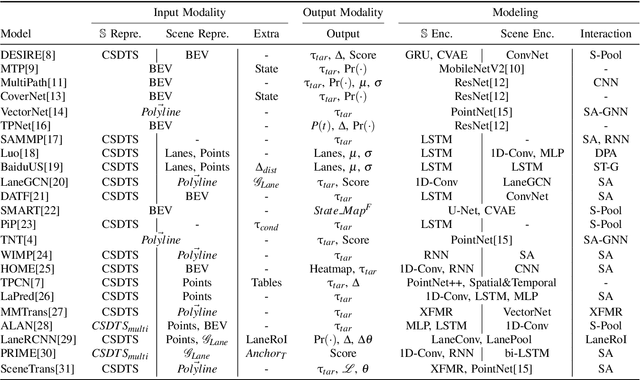
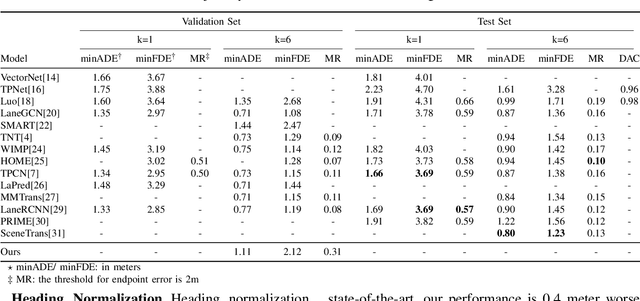
Abstract:With the rapid development of machine learning, autonomous driving has become a hot issue, making urgent demands for more intelligent perception and planning systems. Self-driving cars can avoid traffic crashes with precisely predicted future trajectories of surrounding vehicles. In this work, we review and categorize existing learning-based trajectory forecasting methods from perspectives of representation, modeling, and learning. Moreover, we make our implementation of Target-driveN Trajectory Prediction publicly available at https://github.com/Henry1iu/TNT-Trajectory-Predition, demonstrating its outstanding performance whereas its original codes are withheld. Enlightenment is expected for researchers seeking to improve trajectory prediction performance based on the achievement we have made.
A Large-Scale Dataset for Benchmarking Elevator Button Segmentation and Character Recognition
Mar 22, 2021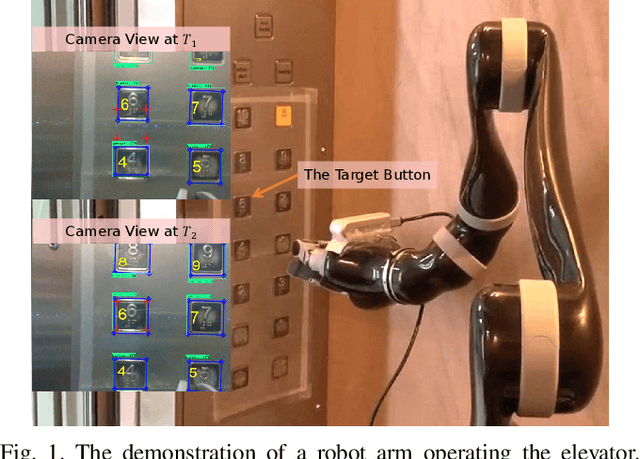
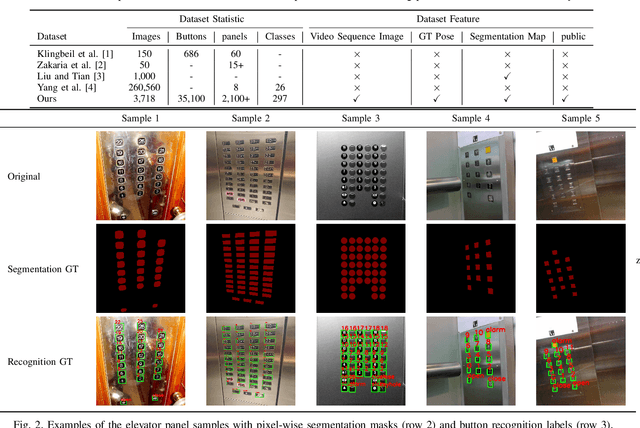
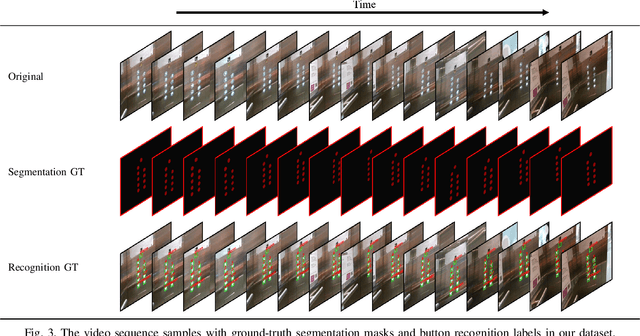
Abstract:Human activities are hugely restricted by COVID-19, recently. Robots that can conduct inter-floor navigation attract much public attention, since they can substitute human workers to conduct the service work. However, current robots either depend on human assistance or elevator retrofitting, and fully autonomous inter-floor navigation is still not available. As the very first step of inter-floor navigation, elevator button segmentation and recognition hold an important position. Therefore, we release the first large-scale publicly available elevator panel dataset in this work, containing 3,718 panel images with 35,100 button labels, to facilitate more powerful algorithms on autonomous elevator operation. Together with the dataset, a number of deep learning based implementations for button segmentation and recognition are also released to benchmark future methods in the community. The dataset will be available at \url{https://github.com/zhudelong/elevator_button_recognition
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge