Shijun Qiu
Source-Free Collaborative Domain Adaptation via Multi-Perspective Feature Enrichment for Functional MRI Analysis
Aug 24, 2023



Abstract:Resting-state functional MRI (rs-fMRI) is increasingly employed in multi-site research to aid neurological disorder analysis. Existing studies usually suffer from significant cross-site/domain data heterogeneity caused by site effects such as differences in scanners/protocols. Many methods have been proposed to reduce fMRI heterogeneity between source and target domains, heavily relying on the availability of source data. But acquiring source data is challenging due to privacy concerns and/or data storage burdens in multi-site studies. To this end, we design a source-free collaborative domain adaptation (SCDA) framework for fMRI analysis, where only a pretrained source model and unlabeled target data are accessible. Specifically, a multi-perspective feature enrichment method (MFE) is developed for target fMRI analysis, consisting of multiple collaborative branches to dynamically capture fMRI features of unlabeled target data from multiple views. Each branch has a data-feeding module, a spatiotemporal feature encoder, and a class predictor. A mutual-consistency constraint is designed to encourage pair-wise consistency of latent features of the same input generated from these branches for robust representation learning. To facilitate efficient cross-domain knowledge transfer without source data, we initialize MFE using parameters of a pretrained source model. We also introduce an unsupervised pretraining strategy using 3,806 unlabeled fMRIs from three large-scale auxiliary databases, aiming to obtain a general feature encoder. Experimental results on three public datasets and one private dataset demonstrate the efficacy of our method in cross-scanner and cross-study prediction tasks. The model pretrained on large-scale rs-fMRI data has been released to the public.
Brain Anatomy Prior Modeling to Forecast Clinical Progression of Cognitive Impairment with Structural MRI
Jun 26, 2023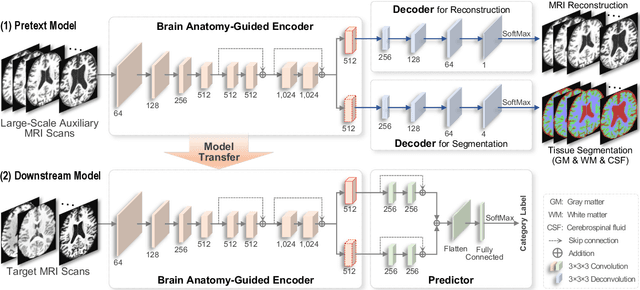
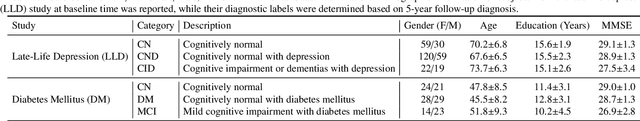
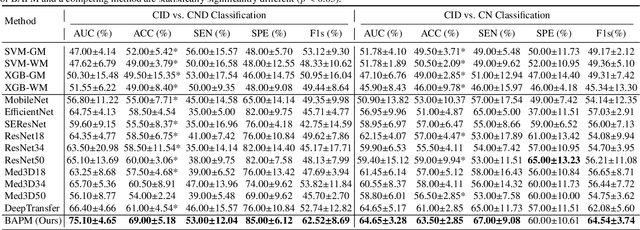
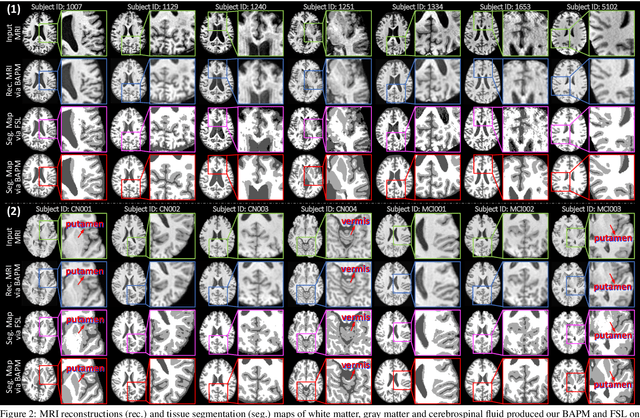
Abstract:Brain structural MRI has been widely used to assess the future progression of cognitive impairment (CI). Previous learning-based studies usually suffer from the issue of small-sized labeled training data, while there exist a huge amount of structural MRIs in large-scale public databases. Intuitively, brain anatomical structures derived from these public MRIs (even without task-specific label information) can be used to boost CI progression trajectory prediction. However, previous studies seldom take advantage of such brain anatomy prior. To this end, this paper proposes a brain anatomy prior modeling (BAPM) framework to forecast the clinical progression of cognitive impairment with small-sized target MRIs by exploring anatomical brain structures. Specifically, the BAPM consists of a pretext model and a downstream model, with a shared brain anatomy-guided encoder to model brain anatomy prior explicitly. Besides the encoder, the pretext model also contains two decoders for two auxiliary tasks (i.e., MRI reconstruction and brain tissue segmentation), while the downstream model relies on a predictor for classification. The brain anatomy-guided encoder is pre-trained with the pretext model on 9,344 auxiliary MRIs without diagnostic labels for anatomy prior modeling. With this encoder frozen, the downstream model is then fine-tuned on limited target MRIs for prediction. We validate the BAPM on two CI-related studies with T1-weighted MRIs from 448 subjects. Experimental results suggest the effectiveness of BAPM in (1) four CI progression prediction tasks, (2) MR image reconstruction, and (3) brain tissue segmentation, compared with several state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge