DeepNoise: Disentanglement of Experimental Noise from Real Biological Signals based on Fluorescent Microscopy Image Classification via Deep Learning
Paper and Code
Sep 13, 2022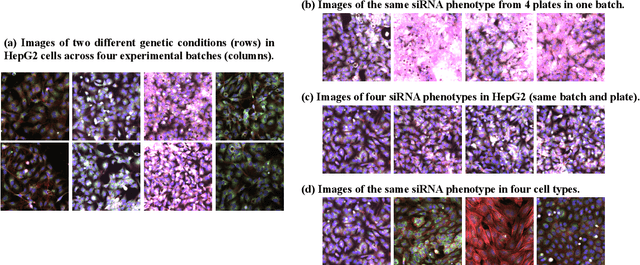
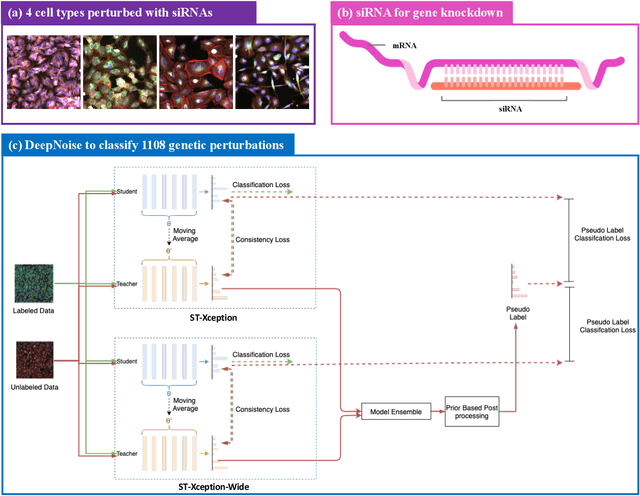
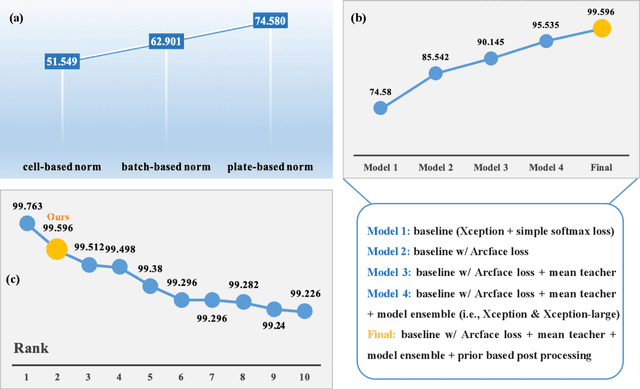
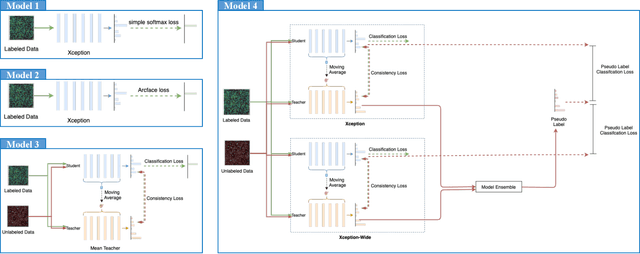
The high-content image-based assay is commonly leveraged for identifying the phenotypic impact of genetic perturbations in biology field. However, a persistent issue remains unsolved during experiments: the interferential technical noise caused by systematic errors (e.g., temperature, reagent concentration, and well location) is always mixed up with the real biological signals, leading to misinterpretation of any conclusion drawn. Here, we show a mean teacher based deep learning model (DeepNoise) that can disentangle biological signals from the experimental noise. Specifically, we aim to classify the phenotypic impact of 1,108 different genetic perturbations screened from 125,510 fluorescent microscopy images, which are totally unrecognizable by human eye. We validate our model by participating in the Recursion Cellular Image Classification Challenge, and our proposed method achieves an extremely high classification score (Acc: 99.596%), ranking the 2nd place among 866 participating groups. This promising result indicates the successful separation of biological and technical factors, which might help decrease the cost of treatment development and expedite the drug discovery process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge