Xiaoping Yang
Senior Member, IEEE
A Fully Automatic Framework for Intracranial Pressure Grading: Integrating Keyframe Identification, ONSD Measurement and Clinical Data
Sep 11, 2025Abstract:Intracranial pressure (ICP) elevation poses severe threats to cerebral function, thus necessitating monitoring for timely intervention. While lumbar puncture is the gold standard for ICP measurement, its invasiveness and associated risks drive the need for non-invasive alternatives. Optic nerve sheath diameter (ONSD) has emerged as a promising biomarker, as elevated ICP directly correlates with increased ONSD. However, current clinical practices for ONSD measurement suffer from inconsistency in manual operation, subjectivity in optimal view selection, and variability in thresholding, limiting their reliability. To address these challenges, we introduce a fully automatic two-stage framework for ICP grading, integrating keyframe identification, ONSD measurement and clinical data. Specifically, the fundus ultrasound video processing stage performs frame-level anatomical segmentation, rule-based keyframe identification guided by an international consensus statement, and precise ONSD measurement. The intracranial pressure grading stage then fuses ONSD metrics with clinical features to enable the prediction of ICP grades, thereby demonstrating an innovative blend of interpretable ultrasound analysis and multi-source data integration for objective clinical evaluation. Experimental results demonstrate that our method achieves a validation accuracy of $0.845 \pm 0.071$ (with standard deviation from five-fold cross-validation) and an independent test accuracy of 0.786, significantly outperforming conventional threshold-based method ($0.637 \pm 0.111$ validation accuracy, $0.429$ test accuracy). Through effectively reducing operator variability and integrating multi-source information, our framework establishes a reliable non-invasive approach for clinical ICP evaluation, holding promise for improving patient management in acute neurological conditions.
A Weakly Supervised Segmentation Network Embedding Cross-scale Attention Guidance and Noise-sensitive Constraint for Detecting Tertiary Lymphoid Structures of Pancreatic Tumors
Jul 27, 2023


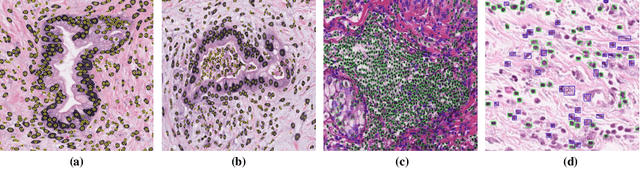
Abstract:The presence of tertiary lymphoid structures (TLSs) on pancreatic pathological images is an important prognostic indicator of pancreatic tumors. Therefore, TLSs detection on pancreatic pathological images plays a crucial role in diagnosis and treatment for patients with pancreatic tumors. However, fully supervised detection algorithms based on deep learning usually require a large number of manual annotations, which is time-consuming and labor-intensive. In this paper, we aim to detect the TLSs in a manner of few-shot learning by proposing a weakly supervised segmentation network. We firstly obtain the lymphocyte density maps by combining a pretrained model for nuclei segmentation and a domain adversarial network for lymphocyte nuclei recognition. Then, we establish a cross-scale attention guidance mechanism by jointly learning the coarse-scale features from the original histopathology images and fine-scale features from our designed lymphocyte density attention. A noise-sensitive constraint is introduced by an embedding signed distance function loss in the training procedure to reduce tiny prediction errors. Experimental results on two collected datasets demonstrate that our proposed method significantly outperforms the state-of-the-art segmentation-based algorithms in terms of TLSs detection accuracy. Additionally, we apply our method to study the congruent relationship between the density of TLSs and peripancreatic vascular invasion and obtain some clinically statistical results.
The optimal connection model for blood vessels segmentation and the MEA-Net
Jun 02, 2023Abstract:Vascular diseases have long been regarded as a significant health concern. Accurately detecting the location, shape, and afflicted regions of blood vessels from a diverse range of medical images has proven to be a major challenge. Obtaining blood vessels that retain their correct topological structures is currently a crucial research issue. Numerous efforts have sought to reinforce neural networks' learning of vascular geometric features, including measures to ensure the correct topological structure of the segmentation result's vessel centerline. Typically, these methods extract topological features from the network's segmentation result and then apply regular constraints to reinforce the accuracy of critical components and the overall topological structure. However, as blood vessels are three-dimensional structures, it is essential to achieve complete local vessel segmentation, which necessitates enhancing the segmentation of vessel boundaries. Furthermore, current methods are limited to handling 2D blood vessel fragmentation cases. Our proposed boundary attention module directly extracts boundary voxels from the network's segmentation result. Additionally, we have established an optimal connection model based on minimal surfaces to determine the connection order between blood vessels. Our method achieves state-of-the-art performance in 3D multi-class vascular segmentation tasks, as evidenced by the high values of Dice Similarity Coefficient (DSC) and Normalized Surface Dice (NSD) metrics. Furthermore, our approach improves the Betti error, LR error, and BR error indicators of vessel richness and structural integrity by more than 10% compared to other methods, and effectively addresses vessel fragmentation and yields blood vessels with a more precise topological structure.
Automated Peripancreatic Vessel Segmentation and Labeling Based on Iterative Trunk Growth and Weakly Supervised Mechanism
Mar 06, 2023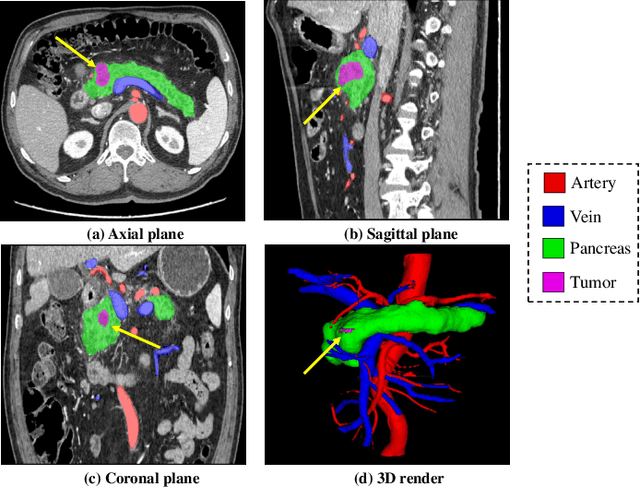
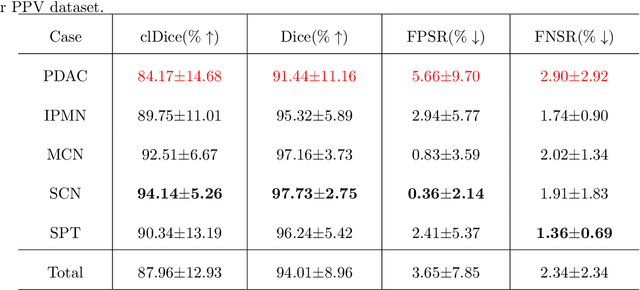
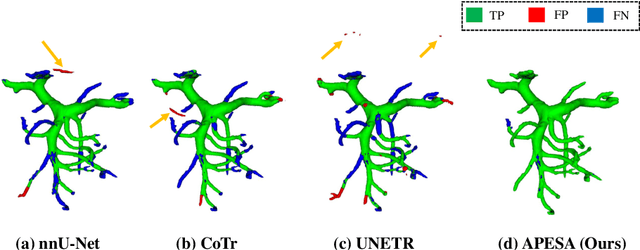
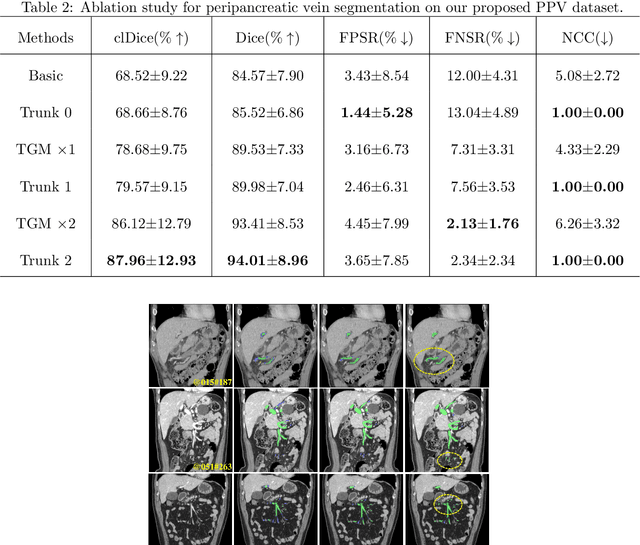
Abstract:Peripancreatic vessel segmentation and anatomical labeling play extremely important roles to assist the early diagnosis, surgery planning and prognosis for patients with pancreatic tumors. However, most current techniques cannot achieve satisfactory segmentation performance for peripancreatic veins and usually make predictions with poor integrity and connectivity. Besides, unsupervised labeling algorithms cannot deal with complex anatomical variation while fully supervised methods require a large number of voxel-wise annotations for training, which is very labor-intensive and time-consuming. To address these problems, we propose our Automated Peripancreatic vEssel Segmentation and lAbeling (APESA) framework, to not only highly improve the segmentation performance for peripancreatic veins, but also efficiently identify the peripancreatic artery branches. There are two core modules in our proposed APESA framework: iterative trunk growth module (ITGM) for vein segmentation and weakly supervised labeling mechanism (WSLM) for artery branch identification. Our proposed ITGM is composed of a series of trunk growth modules, each of which chooses the most reliable trunk of a basic vessel prediction by the largest connected constraint, and seeks for the possible growth branches by branch proposal network. Our designed iterative process guides the raw trunk to be more complete and fully connected. Our proposed WSLM consists of an unsupervised rule-based preprocessing for generating pseudo branch annotations, and an anatomical labeling network to learn the branch distribution voxel by voxel. We achieve Dice of 94.01% for vein segmentation on our collected dataset, which boosts the accuracy by nearly 10% compared with the state-of-the-art methods. Additionally, we also achieve Dice of 97.01% on segmentation and competitive performance on anatomical labeling for peripancreatic arteries.
CTG-Net: An Efficient Cascaded Framework Driven by Terminal Guidance Mechanism for Dilated Pancreatic Duct Segmentation
Mar 06, 2023Abstract:Pancreatic duct dilation indicates a high risk of various pancreatic diseases. Segmentation of dilated pancreatic ducts on computed tomography (CT) images shows the potential to assist the early diagnosis, surgical planning and prognosis. Because of the ducts' tiny sizes, slender tubular structures and the surrounding distractions, most current researches on pancreatic duct segmentation achieve low accuracy and always have segmentation errors on the terminal parts of the ducts. To address these problems, we propose a terminal guidance mechanism called cascaded terminal guidance network (CTG-Net). Firstly, a terminal attention mechanism is established on the skeletons extracted from the coarse predictions. Then, to get fine terminal segmentation, a subnetwork is designed for jointly learning the local intensity from the original images, feature cues from coarse predictions and global anatomy information from the pancreas distance transform maps. Finally, a terminal distraction attention module which explicitly learns the distribution of the terminal distraction is proposed to reduce the false positive and false negative predictions. We also propose a new metric called tDice to measure the terminal segmentation accuracy for targets with tubular structures and two segmentation metrics for distractions. We collect our dilated pancreatic duct segmentation dataset with 150 CT scans from patients with 5 types of pancreatic tumors. Experimental results on our dataset show that our proposed approach boosts dilated pancreatic duct segmentation accuracy by nearly 20% compared with the existing results, and achieves more than 9% improvement for the terminal segmentation accuracy compared with the state-of-the-art methods.
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022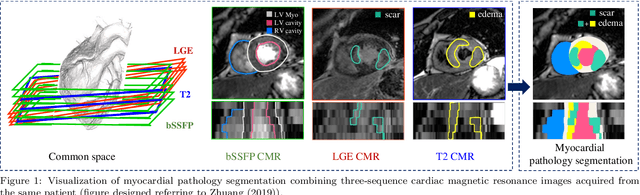
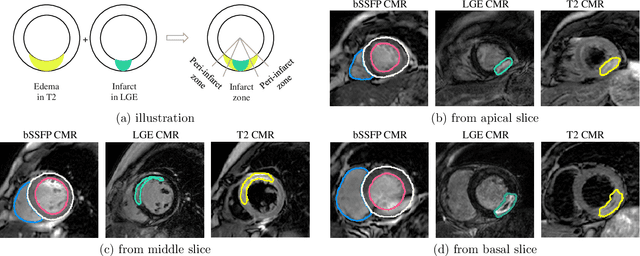
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
Combining CNN and Hybrid Active Contours for Head and Neck Tumor Segmentation in CT and PET images
Dec 28, 2020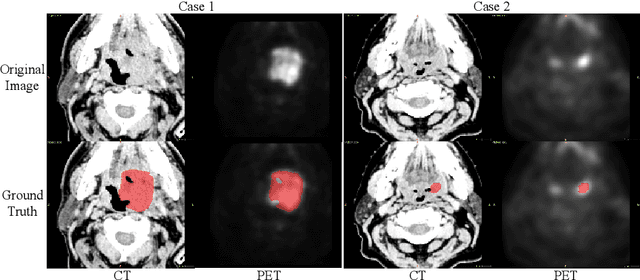
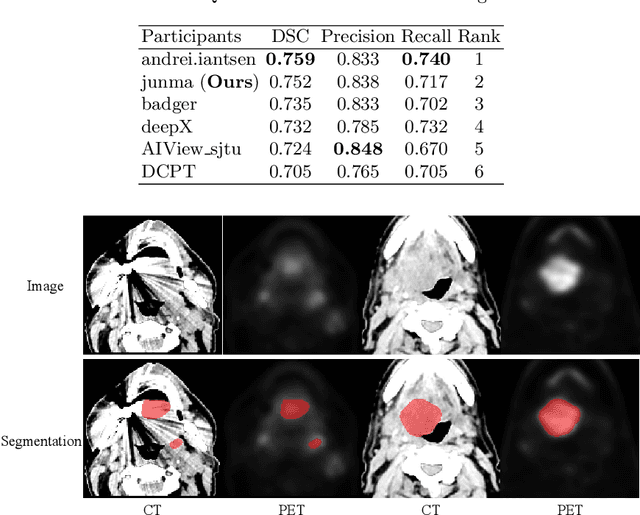
Abstract:Automatic segmentation of head and neck tumors plays an important role in radiomics analysis. In this short paper, we propose an automatic segmentation method for head and neck tumors from PET and CT images based on the combination of convolutional neural networks (CNNs) and hybrid active contours. Specifically, we first introduce a multi-channel 3D U-Net to segment the tumor with the concatenated PET and CT images. Then, we estimate the segmentation uncertainty by model ensembles and define a segmentation quality score to select the cases with high uncertainties. Finally, we develop a hybrid active contour model to refine the high uncertainty cases. Our method ranked second place in the MICCAI 2020 HECKTOR challenge with average Dice Similarity Coefficient, precision, and recall of 0.752, 0.838, and 0.717, respectively.
AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?
Oct 28, 2020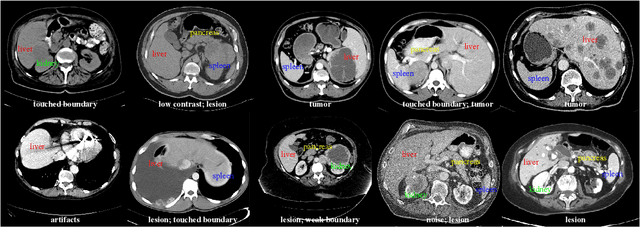
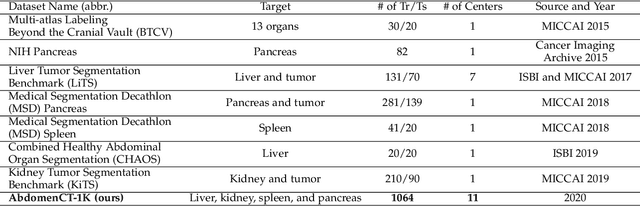
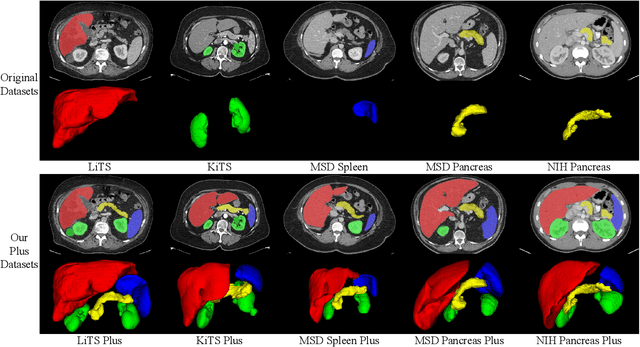
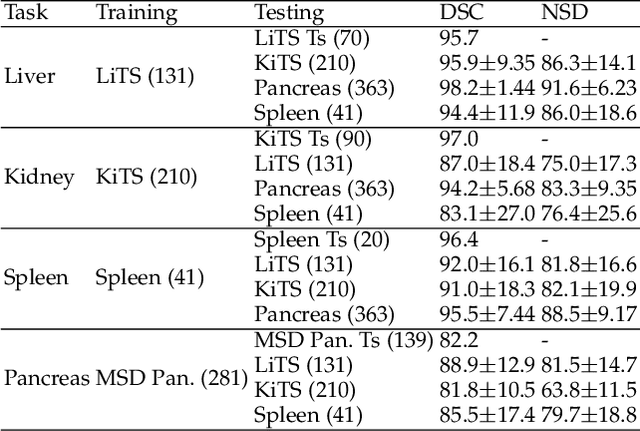
Abstract:With the unprecedented developments in deep learning, automatic segmentation of main abdominal organs (i.e., liver, kidney, and spleen) seems to be a solved problem as the state-of-the-art (SOTA) methods have achieved comparable results with inter-observer variability on existing benchmark datasets. However, most of the existing abdominal organ segmentation benchmark datasets only contain single-center, single-phase, single-vendor, or single-disease cases, thus, it is unclear whether the excellent performance can generalize on more diverse datasets. In this paper, we present a large and diverse abdominal CT organ segmentation dataset, termed as AbdomenCT-1K, with more than 1000 (1K) CT scans from 11 countries, including multi-center, multi-phase, multi-vendor, and multi-disease cases. Furthermore, we conduct a large-scale study for liver, kidney, spleen, and pancreas segmentation, as well as reveal the unsolved segmentation problems of the SOTA method, such as the limited generalization ability on distinct medical centers, phases, and unseen diseases. To advance the unsolved problems, we build four organ segmentation benchmarks for fully supervised, semi-supervised, weakly supervised, and continual learning, which are currently challenging and active research topics. Accordingly, we develop a simple and effective method for each benchmark, which can be used as out-of-the-box methods and strong baselines. We believe the introduction of the AbdomenCT-1K dataset will promote future in-depth research towards clinical applicable abdominal organ segmentation methods. Moreover, the datasets, codes, and trained models of baseline methods will be publicly available at https://github.com/JunMa11/AbdomenCT-1K.
Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Jul 11, 2020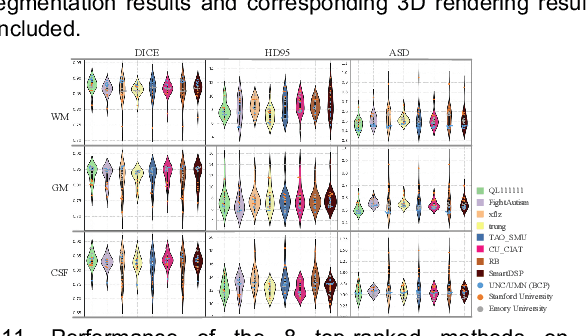
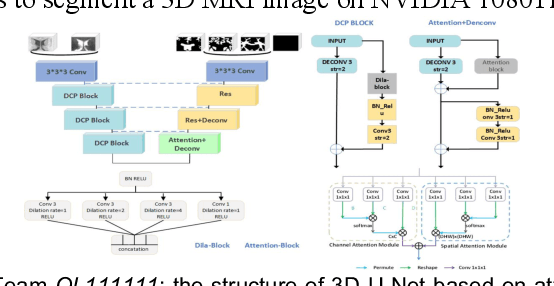
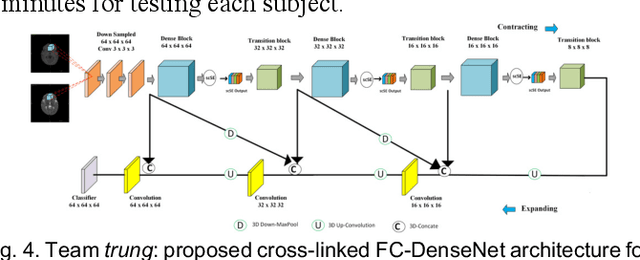
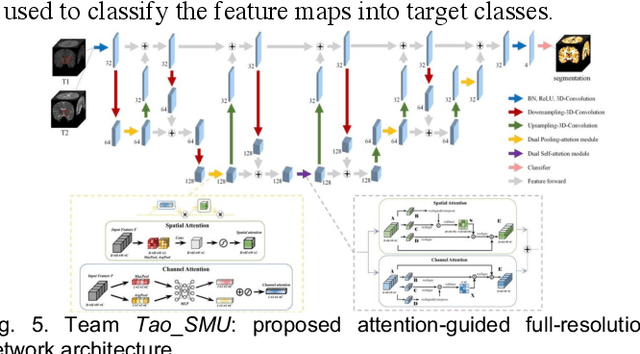
Abstract:To better understand early brain growth patterns in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participating in iSeg-2019. We review the 8 top-ranked teams by detailing their pipelines/implementations, presenting experimental results and evaluating performance in terms of the whole brain, regions of interest, and gyral landmark curves. We also discuss their limitations and possible future directions for the multi-site issue. We hope that the multi-site dataset in iSeg-2019 and this review article will attract more researchers on the multi-site issue.
A Fast Algorithm for Geodesic Active Contours with Applications to Medical Image Segmentation
Jul 01, 2020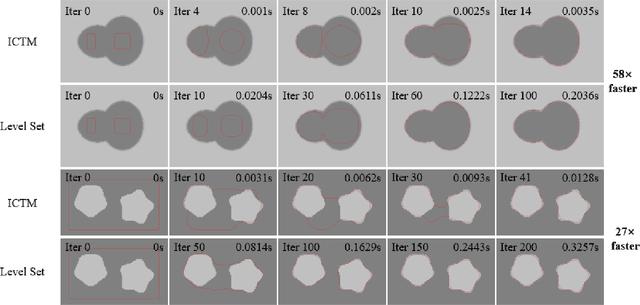
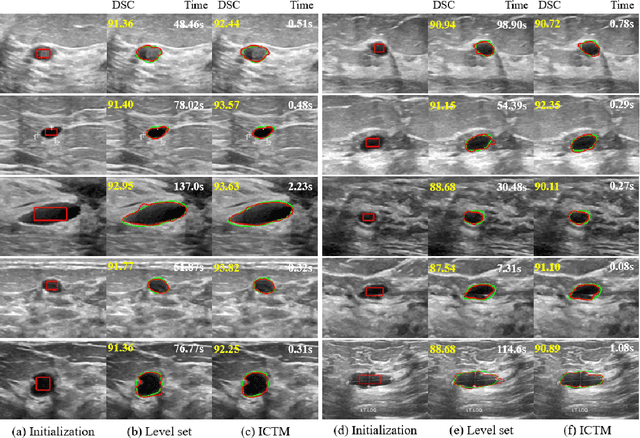
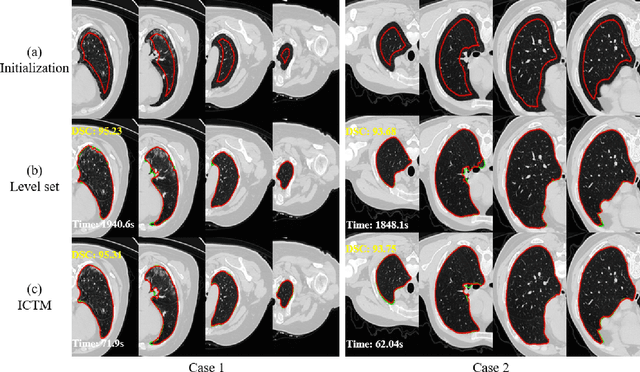
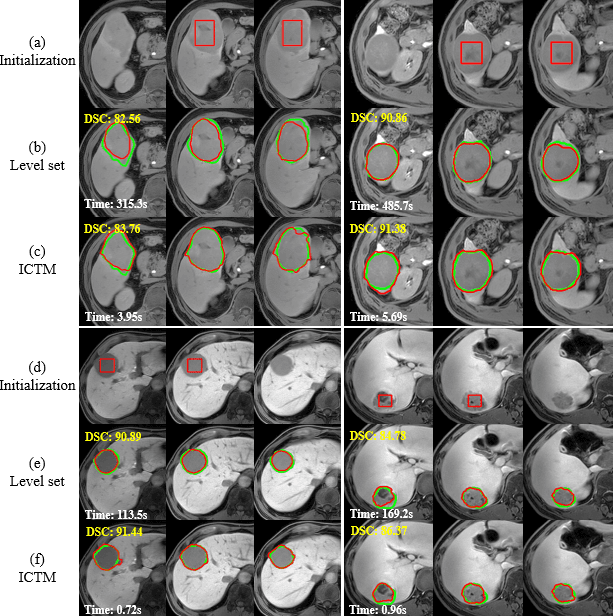
Abstract:The geodesic active contour model (GAC) is a commonly used segmentation model for medical image segmentation. The level set method (LSM) is the most popular approach for solving the model, via implicitly representing the contour by a level set function. However, the LSM suffers from high computation burden and numerical instability, requiring additional regularization terms or re-initialization techniques. In this paper, we use characteristic functions to implicitly approximate the contours, propose a new representation to the GAC and derive an efficient algorithm termed as the iterative convolution-thresholding method (ICTM). Compared to the LSM, the ICTM is simpler and much more efficient and stable. In addition, the ICTM enjoys most desired features (e.g., topological changes) of the level set-based methods. Extensive experiments, on 2D synthetic, 2D ultrasound, 3D CT, and 3D MR images for nodule, organ and lesion segmentation, demonstrate that the ICTM not only obtains comparable or even better segmentation results (compared to the LSM) but also achieves dozens or hundreds of times acceleration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge