Zhenghua Cai
A Weakly Supervised Segmentation Network Embedding Cross-scale Attention Guidance and Noise-sensitive Constraint for Detecting Tertiary Lymphoid Structures of Pancreatic Tumors
Jul 27, 2023


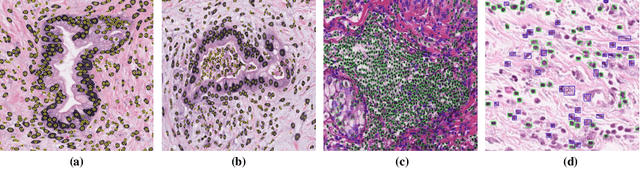
Abstract:The presence of tertiary lymphoid structures (TLSs) on pancreatic pathological images is an important prognostic indicator of pancreatic tumors. Therefore, TLSs detection on pancreatic pathological images plays a crucial role in diagnosis and treatment for patients with pancreatic tumors. However, fully supervised detection algorithms based on deep learning usually require a large number of manual annotations, which is time-consuming and labor-intensive. In this paper, we aim to detect the TLSs in a manner of few-shot learning by proposing a weakly supervised segmentation network. We firstly obtain the lymphocyte density maps by combining a pretrained model for nuclei segmentation and a domain adversarial network for lymphocyte nuclei recognition. Then, we establish a cross-scale attention guidance mechanism by jointly learning the coarse-scale features from the original histopathology images and fine-scale features from our designed lymphocyte density attention. A noise-sensitive constraint is introduced by an embedding signed distance function loss in the training procedure to reduce tiny prediction errors. Experimental results on two collected datasets demonstrate that our proposed method significantly outperforms the state-of-the-art segmentation-based algorithms in terms of TLSs detection accuracy. Additionally, we apply our method to study the congruent relationship between the density of TLSs and peripancreatic vascular invasion and obtain some clinically statistical results.
CTG-Net: An Efficient Cascaded Framework Driven by Terminal Guidance Mechanism for Dilated Pancreatic Duct Segmentation
Mar 06, 2023Abstract:Pancreatic duct dilation indicates a high risk of various pancreatic diseases. Segmentation of dilated pancreatic ducts on computed tomography (CT) images shows the potential to assist the early diagnosis, surgical planning and prognosis. Because of the ducts' tiny sizes, slender tubular structures and the surrounding distractions, most current researches on pancreatic duct segmentation achieve low accuracy and always have segmentation errors on the terminal parts of the ducts. To address these problems, we propose a terminal guidance mechanism called cascaded terminal guidance network (CTG-Net). Firstly, a terminal attention mechanism is established on the skeletons extracted from the coarse predictions. Then, to get fine terminal segmentation, a subnetwork is designed for jointly learning the local intensity from the original images, feature cues from coarse predictions and global anatomy information from the pancreas distance transform maps. Finally, a terminal distraction attention module which explicitly learns the distribution of the terminal distraction is proposed to reduce the false positive and false negative predictions. We also propose a new metric called tDice to measure the terminal segmentation accuracy for targets with tubular structures and two segmentation metrics for distractions. We collect our dilated pancreatic duct segmentation dataset with 150 CT scans from patients with 5 types of pancreatic tumors. Experimental results on our dataset show that our proposed approach boosts dilated pancreatic duct segmentation accuracy by nearly 20% compared with the existing results, and achieves more than 9% improvement for the terminal segmentation accuracy compared with the state-of-the-art methods.
Automated Peripancreatic Vessel Segmentation and Labeling Based on Iterative Trunk Growth and Weakly Supervised Mechanism
Mar 06, 2023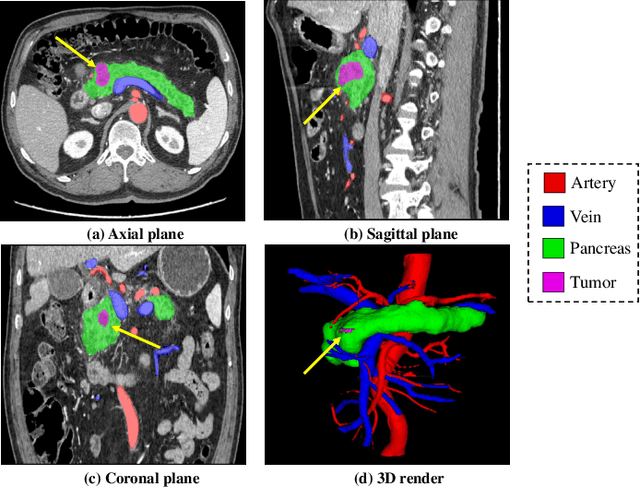
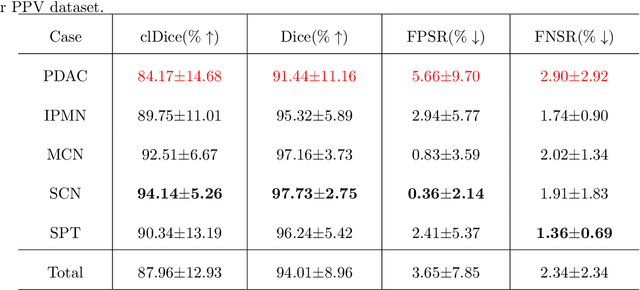
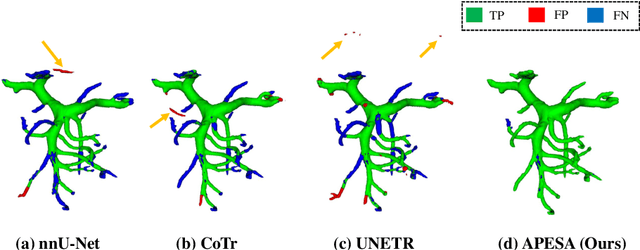
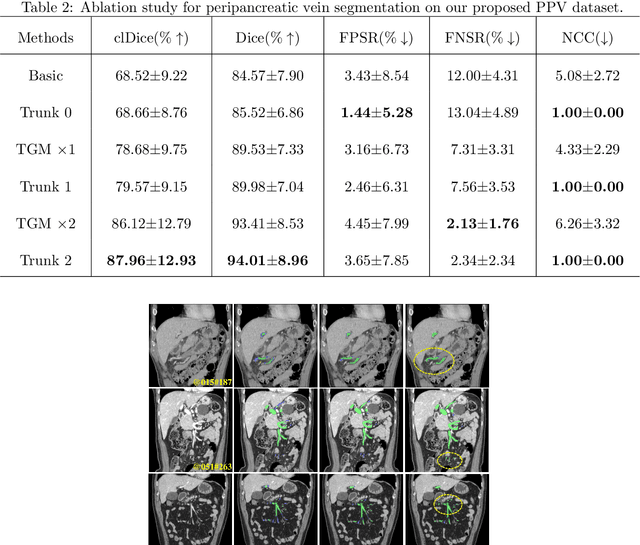
Abstract:Peripancreatic vessel segmentation and anatomical labeling play extremely important roles to assist the early diagnosis, surgery planning and prognosis for patients with pancreatic tumors. However, most current techniques cannot achieve satisfactory segmentation performance for peripancreatic veins and usually make predictions with poor integrity and connectivity. Besides, unsupervised labeling algorithms cannot deal with complex anatomical variation while fully supervised methods require a large number of voxel-wise annotations for training, which is very labor-intensive and time-consuming. To address these problems, we propose our Automated Peripancreatic vEssel Segmentation and lAbeling (APESA) framework, to not only highly improve the segmentation performance for peripancreatic veins, but also efficiently identify the peripancreatic artery branches. There are two core modules in our proposed APESA framework: iterative trunk growth module (ITGM) for vein segmentation and weakly supervised labeling mechanism (WSLM) for artery branch identification. Our proposed ITGM is composed of a series of trunk growth modules, each of which chooses the most reliable trunk of a basic vessel prediction by the largest connected constraint, and seeks for the possible growth branches by branch proposal network. Our designed iterative process guides the raw trunk to be more complete and fully connected. Our proposed WSLM consists of an unsupervised rule-based preprocessing for generating pseudo branch annotations, and an anatomical labeling network to learn the branch distribution voxel by voxel. We achieve Dice of 94.01% for vein segmentation on our collected dataset, which boosts the accuracy by nearly 10% compared with the state-of-the-art methods. Additionally, we also achieve Dice of 97.01% on segmentation and competitive performance on anatomical labeling for peripancreatic arteries.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge