Dorit Merhof
Towards Modality-Agnostic Continual Domain-Incremental Brain Lesion Segmentation
Jan 20, 2026Abstract:Brain lesion segmentation from multi-modal MRI often assumes fixed modality sets or predefined pathologies, making existing models difficult to adapt across cohorts and imaging protocols. Continual learning (CL) offers a natural solution but current approaches either impose a maximum modality configuration or suffer from severe forgetting in buffer-free settings. We introduce CLMU-Net, a replay-based CL framework for 3D brain lesion segmentation that supports arbitrary and variable modality combinations without requiring prior knowledge of the maximum set. A conceptually simple yet effective channel-inflation strategy maps any modality subset into a unified multi-channel representation, enabling a single model to operate across diverse datasets. To enrich inherently local 3D patch features, we incorporate lightweight domain-conditioned textual embeddings that provide global modality-disease context for each training case. Forgetting is further reduced through principled replay using a compact buffer composed of both prototypical and challenging samples. Experiments on five heterogeneous MRI brain datasets demonstrate that CLMU-Net consistently outperforms popular CL baselines. Notably, our method yields an average Dice score improvement of $\geq$ 18\% while remaining robust under heterogeneous-modality conditions. These findings underscore the value of flexible modality handling, targeted replay, and global contextual cues for continual medical image segmentation. Our implementation is available at https://github.com/xmindflow/CLMU-Net.
Tissue Classification and Whole-Slide Images Analysis via Modeling of the Tumor Microenvironment and Biological Pathways
Jan 13, 2026Abstract:Automatic integration of whole slide images (WSIs) and gene expression profiles has demonstrated substantial potential in precision clinical diagnosis and cancer progression studies. However, most existing studies focus on individual gene sequences and slide level classification tasks, with limited attention to spatial transcriptomics and patch level applications. To address this limitation, we propose a multimodal network, BioMorphNet, which automatically integrates tissue morphological features and spatial gene expression to support tissue classification and differential gene analysis. For considering morphological features, BioMorphNet constructs a graph to model the relationships between target patches and their neighbors, and adjusts the response strength based on morphological and molecular level similarity, to better characterize the tumor microenvironment. In terms of multimodal interactions, BioMorphNet derives clinical pathway features from spatial transcriptomic data based on a predefined pathway database, serving as a bridge between tissue morphology and gene expression. In addition, a novel learnable pathway module is designed to automatically simulate the biological pathway formation process, providing a complementary representation to existing clinical pathways. Compared with the latest morphology gene multimodal methods, BioMorphNet's average classification metrics improve by 2.67%, 5.48%, and 6.29% for prostate cancer, colorectal cancer, and breast cancer datasets, respectively. BioMorphNet not only classifies tissue categories within WSIs accurately to support tumor localization, but also analyzes differential gene expression between tissue categories based on prediction confidence, contributing to the discovery of potential tumor biomarkers.
Deep Learning-Based Desikan-Killiany Parcellation of the Brain Using Diffusion MRI
Aug 11, 2025


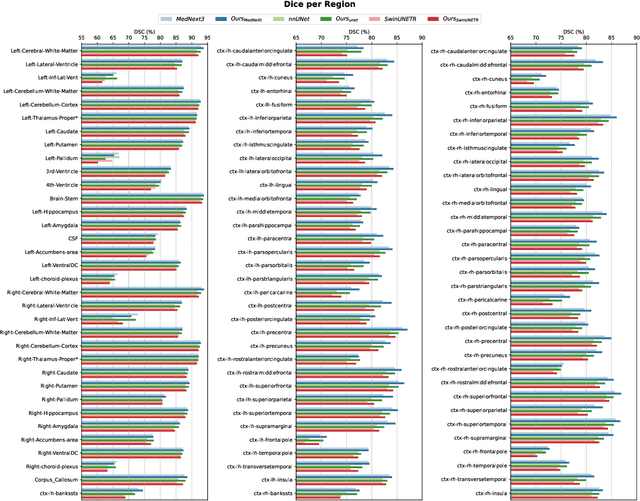
Abstract:Accurate brain parcellation in diffusion MRI (dMRI) space is essential for advanced neuroimaging analyses. However, most existing approaches rely on anatomical MRI for segmentation and inter-modality registration, a process that can introduce errors and limit the versatility of the technique. In this study, we present a novel deep learning-based framework for direct parcellation based on the Desikan-Killiany (DK) atlas using only diffusion MRI data. Our method utilizes a hierarchical, two-stage segmentation network: the first stage performs coarse parcellation into broad brain regions, and the second stage refines the segmentation to delineate more detailed subregions within each coarse category. We conduct an extensive ablation study to evaluate various diffusion-derived parameter maps, identifying an optimal combination of fractional anisotropy, trace, sphericity, and maximum eigenvalue that enhances parellation accuracy. When evaluated on the Human Connectome Project and Consortium for Neuropsychiatric Phenomics datasets, our approach achieves superior Dice Similarity Coefficients compared to existing state-of-the-art models. Additionally, our method demonstrates robust generalization across different image resolutions and acquisition protocols, producing more homogeneous parcellations as measured by the relative standard deviation within regions. This work represents a significant advancement in dMRI-based brain segmentation, providing a precise, reliable, and registration-free solution that is critical for improved structural connectivity and microstructural analyses in both research and clinical applications. The implementation of our method is publicly available on github.com/xmindflow/DKParcellationdMRI.
Spatial Transcriptomics Expression Prediction from Histopathology Based on Cross-Modal Mask Reconstruction and Contrastive Learning
Jun 10, 2025Abstract:Spatial transcriptomics is a technology that captures gene expression levels at different spatial locations, widely used in tumor microenvironment analysis and molecular profiling of histopathology, providing valuable insights into resolving gene expression and clinical diagnosis of cancer. Due to the high cost of data acquisition, large-scale spatial transcriptomics data remain challenging to obtain. In this study, we develop a contrastive learning-based deep learning method to predict spatially resolved gene expression from whole-slide images. Evaluation across six different disease datasets demonstrates that, compared to existing studies, our method improves Pearson Correlation Coefficient (PCC) in the prediction of highly expressed genes, highly variable genes, and marker genes by 6.27%, 6.11%, and 11.26% respectively. Further analysis indicates that our method preserves gene-gene correlations and applies to datasets with limited samples. Additionally, our method exhibits potential in cancer tissue localization based on biomarker expression.
CENet: Context Enhancement Network for Medical Image Segmentation
May 23, 2025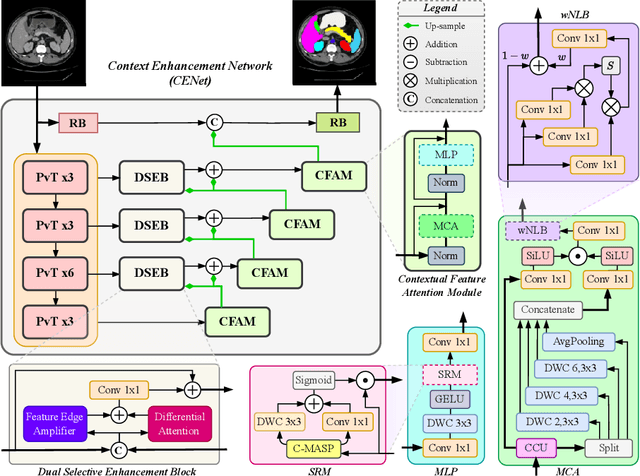
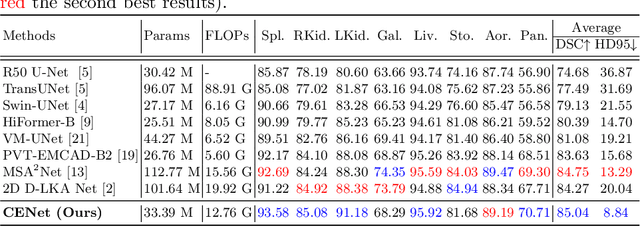
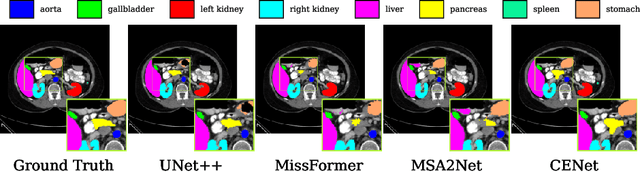
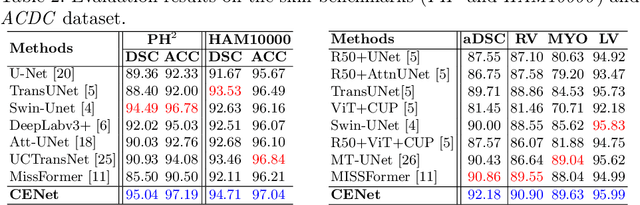
Abstract:Medical image segmentation, particularly in multi-domain scenarios, requires precise preservation of anatomical structures across diverse representations. While deep learning has advanced this field, existing models often struggle with accurate boundary representation, variability in organ morphology, and information loss during downsampling, limiting their accuracy and robustness. To address these challenges, we propose the Context Enhancement Network (CENet), a novel segmentation framework featuring two key innovations. First, the Dual Selective Enhancement Block (DSEB) integrated into skip connections enhances boundary details and improves the detection of smaller organs in a context-aware manner. Second, the Context Feature Attention Module (CFAM) in the decoder employs a multi-scale design to maintain spatial integrity, reduce feature redundancy, and mitigate overly enhanced representations. Extensive evaluations on both radiology and dermoscopic datasets demonstrate that CENet outperforms state-of-the-art (SOTA) methods in multi-organ segmentation and boundary detail preservation, offering a robust and accurate solution for complex medical image analysis tasks. The code is publicly available at https://github.com/xmindflow/cenet.
Attention-based Generative Latent Replay: A Continual Learning Approach for WSI Analysis
May 13, 2025



Abstract:Whole slide image (WSI) classification has emerged as a powerful tool in computational pathology, but remains constrained by domain shifts, e.g., due to different organs, diseases, or institution-specific variations. To address this challenge, we propose an Attention-based Generative Latent Replay Continual Learning framework (AGLR-CL), in a multiple instance learning (MIL) setup for domain incremental WSI classification. Our method employs Gaussian Mixture Models (GMMs) to synthesize WSI representations and patch count distributions, preserving knowledge of past domains without explicitly storing original data. A novel attention-based filtering step focuses on the most salient patch embeddings, ensuring high-quality synthetic samples. This privacy-aware strategy obviates the need for replay buffers and outperforms other buffer-free counterparts while matching the performance of buffer-based solutions. We validate AGLR-CL on clinically relevant biomarker detection and molecular status prediction across multiple public datasets with diverse centers, organs, and patient cohorts. Experimental results confirm its ability to retain prior knowledge and adapt to new domains, offering an effective, privacy-preserving avenue for domain incremental continual learning in WSI classification.
Towards Robust and Generalizable Gerchberg Saxton based Physics Inspired Neural Networks for Computer Generated Holography: A Sensitivity Analysis Framework
Apr 30, 2025Abstract:Computer-generated holography (CGH) enables applications in holographic augmented reality (AR), 3D displays, systems neuroscience, and optical trapping. The fundamental challenge in CGH is solving the inverse problem of phase retrieval from intensity measurements. Physics-inspired neural networks (PINNs), especially Gerchberg-Saxton-based PINNs (GS-PINNs), have advanced phase retrieval capabilities. However, their performance strongly depends on forward models (FMs) and their hyperparameters (FMHs), limiting generalization, complicating benchmarking, and hindering hardware optimization. We present a systematic sensitivity analysis framework based on Saltelli's extension of Sobol's method to quantify FMH impacts on GS-PINN performance. Our analysis demonstrates that SLM pixel-resolution is the primary factor affecting neural network sensitivity, followed by pixel-pitch, propagation distance, and wavelength. Free space propagation forward models demonstrate superior neural network performance compared to Fourier holography, providing enhanced parameterization and generalization. We introduce a composite evaluation metric combining performance consistency, generalization capability, and hyperparameter perturbation resilience, establishing a unified benchmarking standard across CGH configurations. Our research connects physics-inspired deep learning theory with practical CGH implementations through concrete guidelines for forward model selection, neural network architecture, and performance evaluation. Our contributions advance the development of robust, interpretable, and generalizable neural networks for diverse holographic applications, supporting evidence-based decisions in CGH research and implementation.
Modality-Independent Brain Lesion Segmentation with Privacy-aware Continual Learning
Mar 26, 2025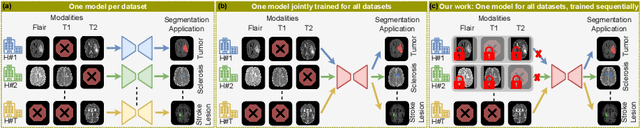
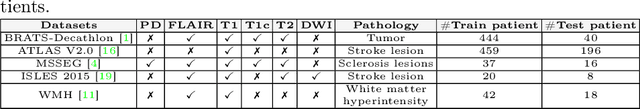
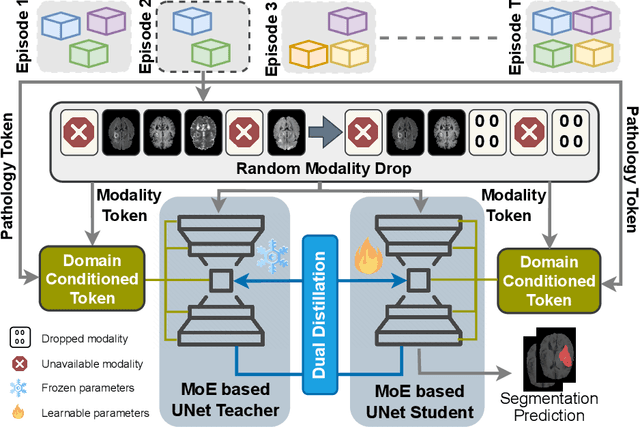
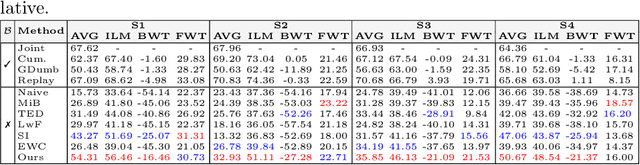
Abstract:Traditional brain lesion segmentation models for multi-modal MRI are typically tailored to specific pathologies, relying on datasets with predefined modalities. Adapting to new MRI modalities or pathologies often requires training separate models, which contrasts with how medical professionals incrementally expand their expertise by learning from diverse datasets over time. Inspired by this human learning process, we propose a unified segmentation model capable of sequentially learning from multiple datasets with varying modalities and pathologies. Our approach leverages a privacy-aware continual learning framework that integrates a mixture-of-experts mechanism and dual knowledge distillation to mitigate catastrophic forgetting while not compromising performance on newly encountered datasets. Extensive experiments across five diverse brain MRI datasets and four dataset sequences demonstrate the effectiveness of our framework in maintaining a single adaptable model, capable of handling varying hospital protocols, imaging modalities, and disease types. Compared to widely used privacy-aware continual learning methods such as LwF, SI, EWC, and MiB, our method achieves an average Dice score improvement of approximately 11%. Our framework represents a significant step toward more versatile and practical brain lesion segmentation models, with implementation available at \href{https://github.com/xmindflow/BrainCL}{GitHub}.
Domain-incremental White Blood Cell Classification with Privacy-aware Continual Learning
Mar 25, 2025Abstract:White blood cell (WBC) classification plays a vital role in hematology for diagnosing various medical conditions. However, it faces significant challenges due to domain shifts caused by variations in sample sources (e.g., blood or bone marrow) and differing imaging conditions across hospitals. Traditional deep learning models often suffer from catastrophic forgetting in such dynamic environments, while foundation models, though generally robust, experience performance degradation when the distribution of inference data differs from that of the training data. To address these challenges, we propose a generative replay-based Continual Learning (CL) strategy designed to prevent forgetting in foundation models for WBC classification. Our method employs lightweight generators to mimic past data with a synthetic latent representation to enable privacy-preserving replay. To showcase the effectiveness, we carry out extensive experiments with a total of four datasets with different task ordering and four backbone models including ResNet50, RetCCL, CTransPath, and UNI. Experimental results demonstrate that conventional fine-tuning methods degrade performance on previously learned tasks and struggle with domain shifts. In contrast, our continual learning strategy effectively mitigates catastrophic forgetting, preserving model performance across varying domains. This work presents a practical solution for maintaining reliable WBC classification in real-world clinical settings, where data distributions frequently evolve.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge