Moein Heidari
Prompt2Perturb (P2P): Text-Guided Diffusion-Based Adversarial Attacks on Breast Ultrasound Images
Dec 13, 2024Abstract:Deep neural networks (DNNs) offer significant promise for improving breast cancer diagnosis in medical imaging. However, these models are highly susceptible to adversarial attacks--small, imperceptible changes that can mislead classifiers--raising critical concerns about their reliability and security. Traditional attacks rely on fixed-norm perturbations, misaligning with human perception. In contrast, diffusion-based attacks require pre-trained models, demanding substantial data when these models are unavailable, limiting practical use in data-scarce scenarios. In medical imaging, however, this is often unfeasible due to the limited availability of datasets. Building on recent advancements in learnable prompts, we propose Prompt2Perturb (P2P), a novel language-guided attack method capable of generating meaningful attack examples driven by text instructions. During the prompt learning phase, our approach leverages learnable prompts within the text encoder to create subtle, yet impactful, perturbations that remain imperceptible while guiding the model towards targeted outcomes. In contrast to current prompt learning-based approaches, our P2P stands out by directly updating text embeddings, avoiding the need for retraining diffusion models. Further, we leverage the finding that optimizing only the early reverse diffusion steps boosts efficiency while ensuring that the generated adversarial examples incorporate subtle noise, thus preserving ultrasound image quality without introducing noticeable artifacts. We show that our method outperforms state-of-the-art attack techniques across three breast ultrasound datasets in FID and LPIPS. Moreover, the generated images are both more natural in appearance and more effective compared to existing adversarial attacks. Our code will be publicly available https://github.com/yasamin-med/P2P.
Single-Layer Learnable Activation for Implicit Neural Representation (SL$^{2}$A-INR)
Sep 18, 2024



Abstract:Implicit Neural Representation (INR), leveraging a neural network to transform coordinate input into corresponding attributes, has recently driven significant advances in several vision-related domains. However, the performance of INR is heavily influenced by the choice of the nonlinear activation function used in its multilayer perceptron (MLP) architecture. Multiple nonlinearities have been investigated; yet, current INRs face limitations in capturing high-frequency components, diverse signal types, and handling inverse problems. We have identified that these problems can be greatly alleviated by introducing a paradigm shift in INRs. We find that an architecture with learnable activations in initial layers can represent fine details in the underlying signals. Specifically, we propose SL$^{2}$A-INR, a hybrid network for INR with a single-layer learnable activation function, prompting the effectiveness of traditional ReLU-based MLPs. Our method performs superior across diverse tasks, including image representation, 3D shape reconstructions, inpainting, single image super-resolution, CT reconstruction, and novel view synthesis. Through comprehensive experiments, SL$^{2}$A-INR sets new benchmarks in accuracy, quality, and convergence rates for INR.
Implicit Neural Representations with Fourier Kolmogorov-Arnold Networks
Sep 14, 2024Abstract:Implicit neural representations (INRs) use neural networks to provide continuous and resolution-independent representations of complex signals with a small number of parameters. However, existing INR models often fail to capture important frequency components specific to each task. To address this issue, in this paper, we propose a Fourier Kolmogorov Arnold network (FKAN) for INRs. The proposed FKAN utilizes learnable activation functions modeled as Fourier series in the first layer to effectively control and learn the task-specific frequency components. In addition, the activation functions with learnable Fourier coefficients improve the ability of the network to capture complex patterns and details, which is beneficial for high-resolution and high-dimensional data. Experimental results show that our proposed FKAN model outperforms three state-of-the-art baseline schemes, and improves the peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM) for the image representation task and intersection over union (IoU) for the 3D occupancy volume representation task, respectively.
MSA$^2$Net: Multi-scale Adaptive Attention-guided Network for Medical Image Segmentation
Aug 03, 2024Abstract:Medical image segmentation involves identifying and separating object instances in a medical image to delineate various tissues and structures, a task complicated by the significant variations in size, shape, and density of these features. Convolutional neural networks (CNNs) have traditionally been used for this task but have limitations in capturing long-range dependencies. Transformers, equipped with self-attention mechanisms, aim to address this problem. However, in medical image segmentation it is beneficial to merge both local and global features to effectively integrate feature maps across various scales, capturing both detailed features and broader semantic elements for dealing with variations in structures. In this paper, we introduce MSA$^2$Net, a new deep segmentation framework featuring an expedient design of skip-connections. These connections facilitate feature fusion by dynamically weighting and combining coarse-grained encoder features with fine-grained decoder feature maps. Specifically, we propose a Multi-Scale Adaptive Spatial Attention Gate (MASAG), which dynamically adjusts the receptive field (Local and Global contextual information) to ensure that spatially relevant features are selectively highlighted while minimizing background distractions. Extensive evaluations involving dermatology, and radiological datasets demonstrate that our MSA$^2$Net outperforms state-of-the-art (SOTA) works or matches their performance. The source code is publicly available at https://github.com/xmindflow/MSA-2Net.
MSA2Net: Multi-scale Adaptive Attention-guided Network for Medical Image Segmentation
Jul 31, 2024Abstract:Medical image segmentation involves identifying and separating object instances in a medical image to delineate various tissues and structures, a task complicated by the significant variations in size, shape, and density of these features. Convolutional neural networks (CNNs) have traditionally been used for this task but have limitations in capturing long-range dependencies. Transformers, equipped with self-attention mechanisms, aim to address this problem. However, in medical image segmentation it is beneficial to merge both local and global features to effectively integrate feature maps across various scales, capturing both detailed features and broader semantic elements for dealing with variations in structures. In this paper, we introduce MSA2Net, a new deep segmentation framework featuring an expedient design of skip-connections. These connections facilitate feature fusion by dynamically weighting and combining coarse-grained encoder features with fine-grained decoder feature maps. Specifically, we propose a Multi-Scale Adaptive Spatial Attention Gate (MASAG), which dynamically adjusts the receptive field (Local and Global contextual information) to ensure that spatially relevant features are selectively highlighted while minimizing background distractions. Extensive evaluations involving dermatology, and radiological datasets demonstrate that our MSA2Net outperforms state-of-the-art (SOTA) works or matches their performance. The source code is publicly available at https://github.com/xmindflow/MSA-2Net.
Computation-Efficient Era: A Comprehensive Survey of State Space Models in Medical Image Analysis
Jun 05, 2024



Abstract:Sequence modeling plays a vital role across various domains, with recurrent neural networks being historically the predominant method of performing these tasks. However, the emergence of transformers has altered this paradigm due to their superior performance. Built upon these advances, transformers have conjoined CNNs as two leading foundational models for learning visual representations. However, transformers are hindered by the $\mathcal{O}(N^2)$ complexity of their attention mechanisms, while CNNs lack global receptive fields and dynamic weight allocation. State Space Models (SSMs), specifically the \textit{\textbf{Mamba}} model with selection mechanisms and hardware-aware architecture, have garnered immense interest lately in sequential modeling and visual representation learning, challenging the dominance of transformers by providing infinite context lengths and offering substantial efficiency maintaining linear complexity in the input sequence. Capitalizing on the advances in computer vision, medical imaging has heralded a new epoch with Mamba models. Intending to help researchers navigate the surge, this survey seeks to offer an encyclopedic review of Mamba models in medical imaging. Specifically, we start with a comprehensive theoretical review forming the basis of SSMs, including Mamba architecture and its alternatives for sequence modeling paradigms in this context. Next, we offer a structured classification of Mamba models in the medical field and introduce a diverse categorization scheme based on their application, imaging modalities, and targeted organs. Finally, we summarize key challenges, discuss different future research directions of the SSMs in the medical domain, and propose several directions to fulfill the demands of this field. In addition, we have compiled the studies discussed in this paper along with their open-source implementations on our GitHub repository.
Vision-Language Synthetic Data Enhances Echocardiography Downstream Tasks
Mar 28, 2024



Abstract:High-quality, large-scale data is essential for robust deep learning models in medical applications, particularly ultrasound image analysis. Diffusion models facilitate high-fidelity medical image generation, reducing the costs associated with acquiring and annotating new images. This paper utilizes recent vision-language models to produce diverse and realistic synthetic echocardiography image data, preserving key features of the original images guided by textual and semantic label maps. Specifically, we investigate three potential avenues: unconditional generation, generation guided by text, and a hybrid approach incorporating both textual and semantic supervision. We show that the rich contextual information present in the synthesized data potentially enhances the accuracy and interpretability of downstream tasks, such as echocardiography segmentation and classification with improved metrics and faster convergence. Our implementation with checkpoints, prompts, and the created synthetic dataset will be publicly available at \href{https://github.com/Pooria90/DiffEcho}{GitHub}.
Enhancing Efficiency in Vision Transformer Networks: Design Techniques and Insights
Mar 28, 2024



Abstract:Intrigued by the inherent ability of the human visual system to identify salient regions in complex scenes, attention mechanisms have been seamlessly integrated into various Computer Vision (CV) tasks. Building upon this paradigm, Vision Transformer (ViT) networks exploit attention mechanisms for improved efficiency. This review navigates the landscape of redesigned attention mechanisms within ViTs, aiming to enhance their performance. This paper provides a comprehensive exploration of techniques and insights for designing attention mechanisms, systematically reviewing recent literature in the field of CV. This survey begins with an introduction to the theoretical foundations and fundamental concepts underlying attention mechanisms. We then present a systematic taxonomy of various attention mechanisms within ViTs, employing redesigned approaches. A multi-perspective categorization is proposed based on their application, objectives, and the type of attention applied. The analysis includes an exploration of the novelty, strengths, weaknesses, and an in-depth evaluation of the different proposed strategies. This culminates in the development of taxonomies that highlight key properties and contributions. Finally, we gather the reviewed studies along with their available open-source implementations at our \href{https://github.com/mindflow-institue/Awesome-Attention-Mechanism-in-Medical-Imaging}{GitHub}\footnote{\url{https://github.com/xmindflow/Awesome-Attention-Mechanism-in-Medical-Imaging}}. We aim to regularly update it with the most recent relevant papers.
SA2-Net: Scale-aware Attention Network for Microscopic Image Segmentation
Sep 28, 2023Abstract:Microscopic image segmentation is a challenging task, wherein the objective is to assign semantic labels to each pixel in a given microscopic image. While convolutional neural networks (CNNs) form the foundation of many existing frameworks, they often struggle to explicitly capture long-range dependencies. Although transformers were initially devised to address this issue using self-attention, it has been proven that both local and global features are crucial for addressing diverse challenges in microscopic images, including variations in shape, size, appearance, and target region density. In this paper, we introduce SA2-Net, an attention-guided method that leverages multi-scale feature learning to effectively handle diverse structures within microscopic images. Specifically, we propose scale-aware attention (SA2) module designed to capture inherent variations in scales and shapes of microscopic regions, such as cells, for accurate segmentation. This module incorporates local attention at each level of multi-stage features, as well as global attention across multiple resolutions. Furthermore, we address the issue of blurred region boundaries (e.g., cell boundaries) by introducing a novel upsampling strategy called the Adaptive Up-Attention (AuA) module. This module enhances the discriminative ability for improved localization of microscopic regions using an explicit attention mechanism. Extensive experiments on five challenging datasets demonstrate the benefits of our SA2-Net model. Our source code is publicly available at \url{https://github.com/mustansarfiaz/SA2-Net}.
Advances in Medical Image Analysis with Vision Transformers: A Comprehensive Review
Jan 10, 2023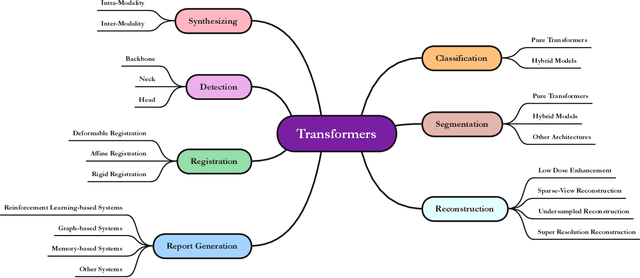
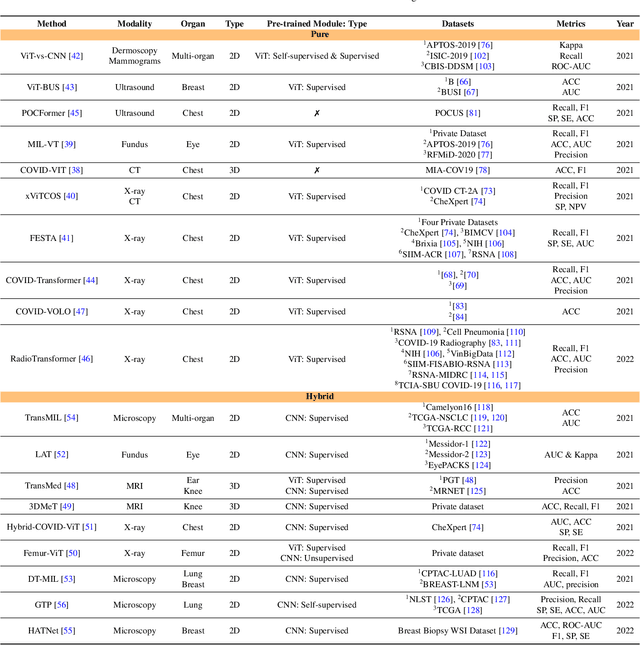
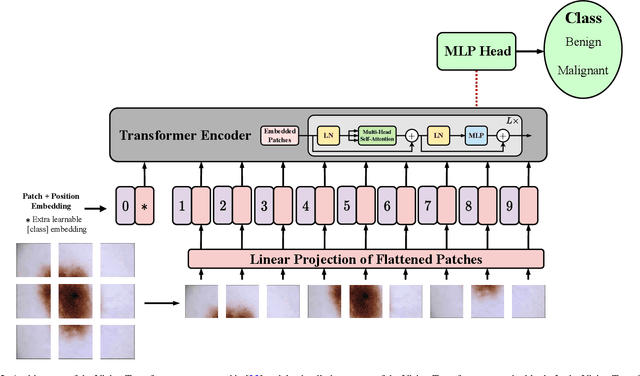
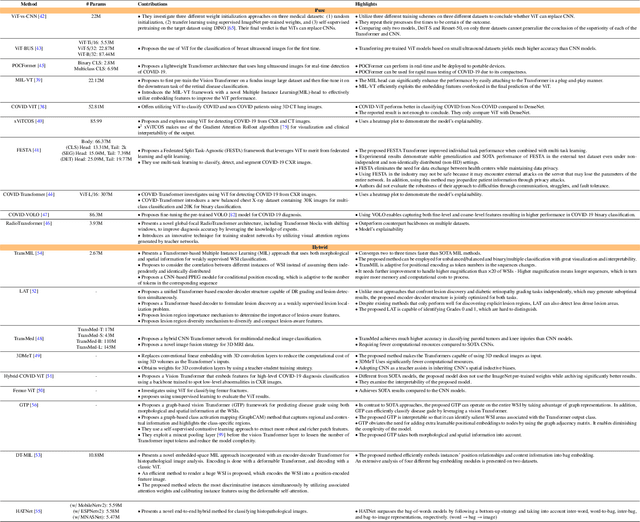
Abstract:The remarkable performance of the Transformer architecture in natural language processing has recently also triggered broad interest in Computer Vision. Among other merits, Transformers are witnessed as capable of learning long-range dependencies and spatial correlations, which is a clear advantage over convolutional neural networks (CNNs), which have been the de facto standard in Computer Vision problems so far. Thus, Transformers have become an integral part of modern medical image analysis. In this review, we provide an encyclopedic review of the applications of Transformers in medical imaging. Specifically, we present a systematic and thorough review of relevant recent Transformer literature for different medical image analysis tasks, including classification, segmentation, detection, registration, synthesis, and clinical report generation. For each of these applications, we investigate the novelty, strengths and weaknesses of the different proposed strategies and develop taxonomies highlighting key properties and contributions. Further, if applicable, we outline current benchmarks on different datasets. Finally, we summarize key challenges and discuss different future research directions. In addition, we have provided cited papers with their corresponding implementations in https://github.com/mindflow-institue/Awesome-Transformer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge