Yousef Sadegheih
Towards Modality-Agnostic Continual Domain-Incremental Brain Lesion Segmentation
Jan 20, 2026Abstract:Brain lesion segmentation from multi-modal MRI often assumes fixed modality sets or predefined pathologies, making existing models difficult to adapt across cohorts and imaging protocols. Continual learning (CL) offers a natural solution but current approaches either impose a maximum modality configuration or suffer from severe forgetting in buffer-free settings. We introduce CLMU-Net, a replay-based CL framework for 3D brain lesion segmentation that supports arbitrary and variable modality combinations without requiring prior knowledge of the maximum set. A conceptually simple yet effective channel-inflation strategy maps any modality subset into a unified multi-channel representation, enabling a single model to operate across diverse datasets. To enrich inherently local 3D patch features, we incorporate lightweight domain-conditioned textual embeddings that provide global modality-disease context for each training case. Forgetting is further reduced through principled replay using a compact buffer composed of both prototypical and challenging samples. Experiments on five heterogeneous MRI brain datasets demonstrate that CLMU-Net consistently outperforms popular CL baselines. Notably, our method yields an average Dice score improvement of $\geq$ 18\% while remaining robust under heterogeneous-modality conditions. These findings underscore the value of flexible modality handling, targeted replay, and global contextual cues for continual medical image segmentation. Our implementation is available at https://github.com/xmindflow/CLMU-Net.
Deep Learning-Based Desikan-Killiany Parcellation of the Brain Using Diffusion MRI
Aug 11, 2025


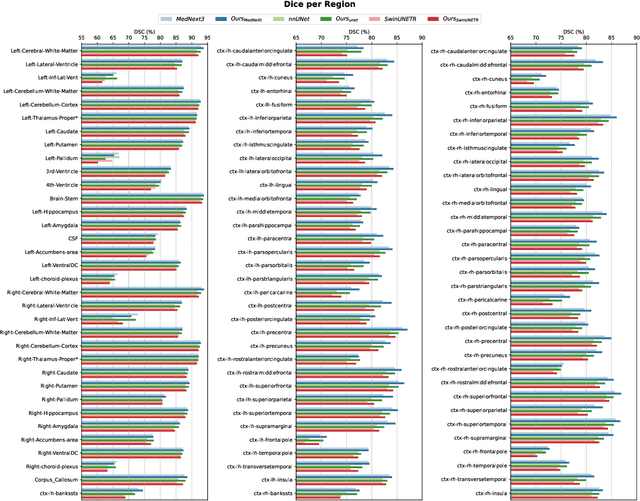
Abstract:Accurate brain parcellation in diffusion MRI (dMRI) space is essential for advanced neuroimaging analyses. However, most existing approaches rely on anatomical MRI for segmentation and inter-modality registration, a process that can introduce errors and limit the versatility of the technique. In this study, we present a novel deep learning-based framework for direct parcellation based on the Desikan-Killiany (DK) atlas using only diffusion MRI data. Our method utilizes a hierarchical, two-stage segmentation network: the first stage performs coarse parcellation into broad brain regions, and the second stage refines the segmentation to delineate more detailed subregions within each coarse category. We conduct an extensive ablation study to evaluate various diffusion-derived parameter maps, identifying an optimal combination of fractional anisotropy, trace, sphericity, and maximum eigenvalue that enhances parellation accuracy. When evaluated on the Human Connectome Project and Consortium for Neuropsychiatric Phenomics datasets, our approach achieves superior Dice Similarity Coefficients compared to existing state-of-the-art models. Additionally, our method demonstrates robust generalization across different image resolutions and acquisition protocols, producing more homogeneous parcellations as measured by the relative standard deviation within regions. This work represents a significant advancement in dMRI-based brain segmentation, providing a precise, reliable, and registration-free solution that is critical for improved structural connectivity and microstructural analyses in both research and clinical applications. The implementation of our method is publicly available on github.com/xmindflow/DKParcellationdMRI.
Modality-Independent Brain Lesion Segmentation with Privacy-aware Continual Learning
Mar 26, 2025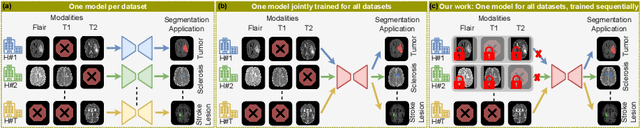
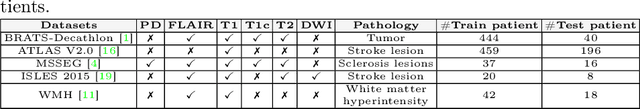
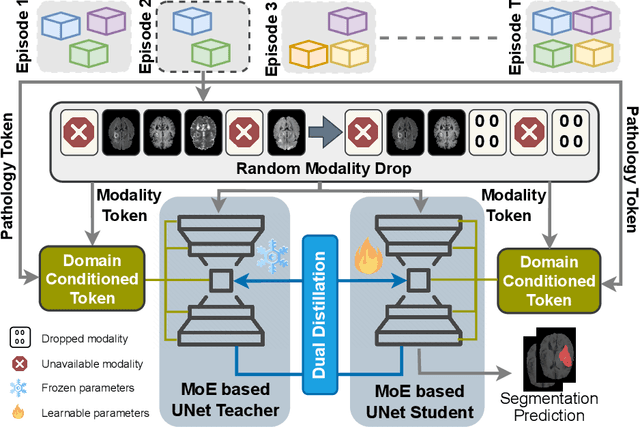
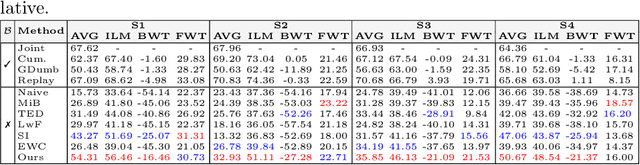
Abstract:Traditional brain lesion segmentation models for multi-modal MRI are typically tailored to specific pathologies, relying on datasets with predefined modalities. Adapting to new MRI modalities or pathologies often requires training separate models, which contrasts with how medical professionals incrementally expand their expertise by learning from diverse datasets over time. Inspired by this human learning process, we propose a unified segmentation model capable of sequentially learning from multiple datasets with varying modalities and pathologies. Our approach leverages a privacy-aware continual learning framework that integrates a mixture-of-experts mechanism and dual knowledge distillation to mitigate catastrophic forgetting while not compromising performance on newly encountered datasets. Extensive experiments across five diverse brain MRI datasets and four dataset sequences demonstrate the effectiveness of our framework in maintaining a single adaptable model, capable of handling varying hospital protocols, imaging modalities, and disease types. Compared to widely used privacy-aware continual learning methods such as LwF, SI, EWC, and MiB, our method achieves an average Dice score improvement of approximately 11%. Our framework represents a significant step toward more versatile and practical brain lesion segmentation models, with implementation available at \href{https://github.com/xmindflow/BrainCL}{GitHub}.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
Segmentation of Brain Metastases in MRI: A Two-Stage Deep Learning Approach with Modality Impact Study
Jul 19, 2024Abstract:Brain metastasis segmentation poses a significant challenge in medical imaging due to the complex presentation and variability in size and location of metastases. In this study, we first investigate the impact of different imaging modalities on segmentation performance using a 3D U-Net. Through a comprehensive analysis, we determine that combining all available modalities does not necessarily enhance performance. Instead, the combination of T1-weighted with contrast enhancement (T1c), T1-weighted (T1), and FLAIR modalities yields superior results. Building on these findings, we propose a two-stage detection and segmentation model specifically designed to accurately segment brain metastases. Our approach demonstrates that leveraging three key modalities (T1c, T1, and FLAIR) achieves significantly higher accuracy compared to single-pass deep learning models. This targeted combination allows for precise segmentation, capturing even small metastases that other models often miss. Our model sets a new benchmark in brain metastasis segmentation, highlighting the importance of strategic modality selection and multi-stage processing in medical imaging. Our implementation is freely accessible to the research community on \href{https://github.com/xmindflow/Met-Seg}{GitHub}.
LHU-Net: A Light Hybrid U-Net for Cost-Efficient, High-Performance Volumetric Medical Image Segmentation
Apr 07, 2024Abstract:As a result of the rise of Transformer architectures in medical image analysis, specifically in the domain of medical image segmentation, a multitude of hybrid models have been created that merge the advantages of Convolutional Neural Networks (CNNs) and Transformers. These hybrid models have achieved notable success by significantly improving segmentation accuracy. Yet, this progress often comes at the cost of increased model complexity, both in terms of parameters and computational demand. Moreover, many of these models fail to consider the crucial interplay between spatial and channel features, which could further refine and improve segmentation outcomes. To address this, we introduce LHU-Net, a Light Hybrid U-Net architecture optimized for volumetric medical image segmentation. LHU-Net is meticulously designed to prioritize spatial feature analysis in its initial layers before shifting focus to channel-based features in its deeper layers, ensuring a comprehensive feature extraction process. Rigorous evaluation across five benchmark datasets - Synapse, LA, Pancreas, ACDC, and BRaTS 2018 - underscores LHU-Net's superior performance, showcasing its dual capacity for efficiency and accuracy. Notably, LHU-Net sets new performance benchmarks, such as attaining a Dice score of 92.66 on the ACDC dataset, while simultaneously reducing parameters by 85% and quartering the computational load compared to existing state-of-the-art models. Achieved without any reliance on pre-training, additional data, or model ensemble, LHU-Net's effectiveness is further evidenced by its state-of-the-art performance across all evaluated datasets, utilizing fewer than 11 million parameters. This achievement highlights that balancing computational efficiency with high accuracy in medical image segmentation is feasible. Our implementation of LHU-Net is freely accessible to the research community on GitHub.
DermoSegDiff: A Boundary-aware Segmentation Diffusion Model for Skin Lesion Delineation
Aug 05, 2023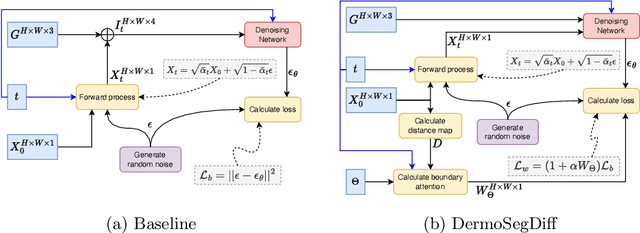


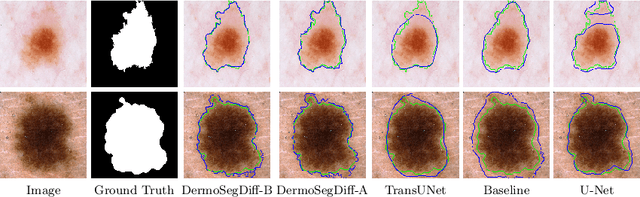
Abstract:Skin lesion segmentation plays a critical role in the early detection and accurate diagnosis of dermatological conditions. Denoising Diffusion Probabilistic Models (DDPMs) have recently gained attention for their exceptional image-generation capabilities. Building on these advancements, we propose DermoSegDiff, a novel framework for skin lesion segmentation that incorporates boundary information during the learning process. Our approach introduces a novel loss function that prioritizes the boundaries during training, gradually reducing the significance of other regions. We also introduce a novel U-Net-based denoising network that proficiently integrates noise and semantic information inside the network. Experimental results on multiple skin segmentation datasets demonstrate the superiority of DermoSegDiff over existing CNN, transformer, and diffusion-based approaches, showcasing its effectiveness and generalization in various scenarios. The implementation is publicly accessible on \href{https://github.com/mindflow-institue/dermosegdiff}{GitHub}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge