Fabian Kiessling
Tissue Concepts: supervised foundation models in computational pathology
Sep 05, 2024



Abstract:Due to the increasing workload of pathologists, the need for automation to support diagnostic tasks and quantitative biomarker evaluation is becoming more and more apparent. Foundation models have the potential to improve generalizability within and across centers and serve as starting points for data efficient development of specialized yet robust AI models. However, the training foundation models themselves is usually very expensive in terms of data, computation, and time. This paper proposes a supervised training method that drastically reduces these expenses. The proposed method is based on multi-task learning to train a joint encoder, by combining 16 different classification, segmentation, and detection tasks on a total of 912,000 patches. Since the encoder is capable of capturing the properties of the samples, we term it the Tissue Concepts encoder. To evaluate the performance and generalizability of the Tissue Concepts encoder across centers, classification of whole slide images from four of the most prevalent solid cancers - breast, colon, lung, and prostate - was used. The experiments show that the Tissue Concepts model achieve comparable performance to models trained with self-supervision, while requiring only 6% of the amount of training patches. Furthermore, the Tissue Concepts encoder outperforms an ImageNet pre-trained encoder on both in-domain and out-of-domain data.
Overcoming Data Scarcity in Biomedical Imaging with a Foundational Multi-Task Model
Nov 16, 2023Abstract:Foundational models, pretrained on a large scale, have demonstrated substantial success across non-medical domains. However, training these models typically requires large, comprehensive datasets, which contrasts with the smaller and more heterogeneous datasets common in biomedical imaging. Here, we propose a multi-task learning strategy that decouples the number of training tasks from memory requirements. We trained a Universal bioMedical PreTrained model (UMedPT) on a multi-task database including tomographic, microscopic, and X-ray images, with various labelling strategies such as classification, segmentation, and object detection. The UMedPT foundational model outperformed ImageNet pretraining and the previous state-of-the-art models. For tasks related to the pretraining database, it maintained its performance with only 1% of the original training data and without fine-tuning. For out-of-domain tasks it required not more than 50% of the original training data. In an external independent validation imaging features extracted using UMedPT proved to be a new standard for cross-center transferability.
Predicting Osteoarthritis Progression in Radiographs via Unsupervised Representation Learning
Nov 22, 2021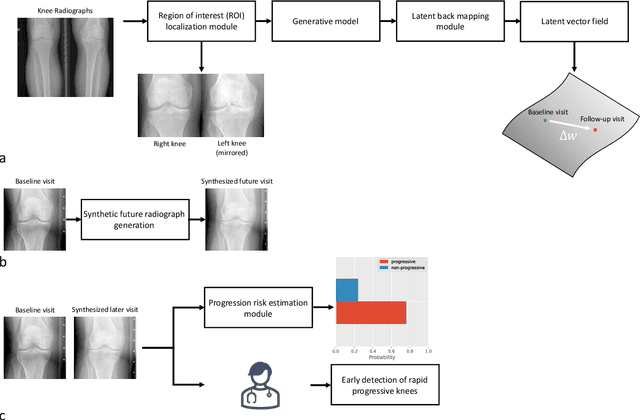
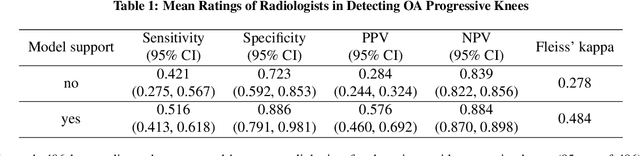
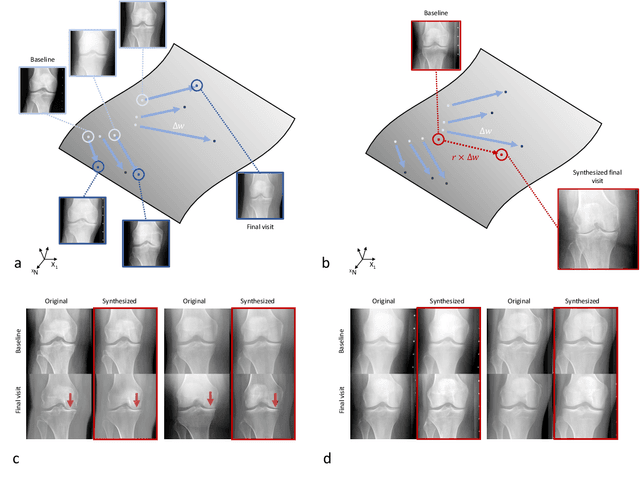
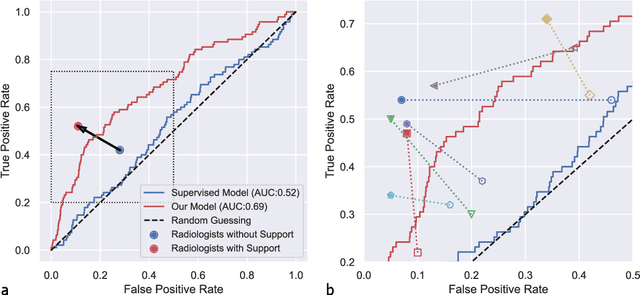
Abstract:Osteoarthritis (OA) is the most common joint disorder affecting substantial proportions of the global population, primarily the elderly. Despite its individual and socioeconomic burden, the onset and progression of OA can still not be reliably predicted. Aiming to fill this diagnostic gap, we introduce an unsupervised learning scheme based on generative models to predict the future development of OA based on knee joint radiographs. Using longitudinal data from osteoarthritis studies, we explore the latent temporal trajectory to predict a patient's future radiographs up to the eight-year follow-up visit. Our model predicts the risk of progression towards OA and surpasses its supervised counterpart whose input was provided by seven experienced radiologists. With the support of the model, sensitivity, specificity, positive predictive value, and negative predictive value increased significantly from 42.1% to 51.6%, from 72.3% to 88.6%, from 28.4% to 57.6%, and from 83.9% to 88.4%, respectively, while without such support, radiologists performed only slightly better than random guessing. Our predictive model improves predictions on OA onset and progression, despite requiring no human annotation in the training phase.
Advancing diagnostic performance and clinical usability of neural networks via adversarial training and dual batch normalization
Nov 25, 2020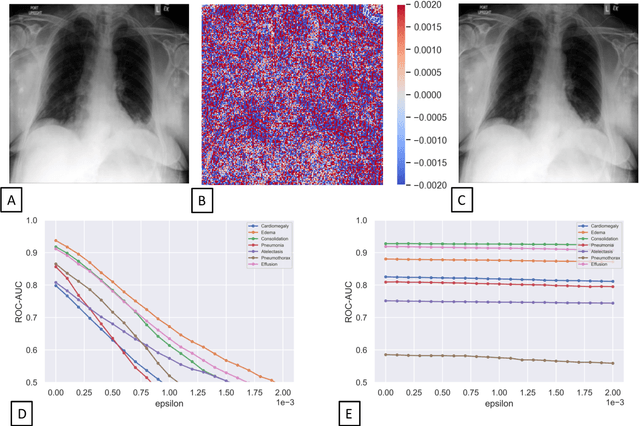
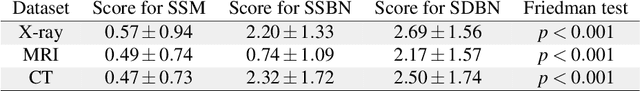
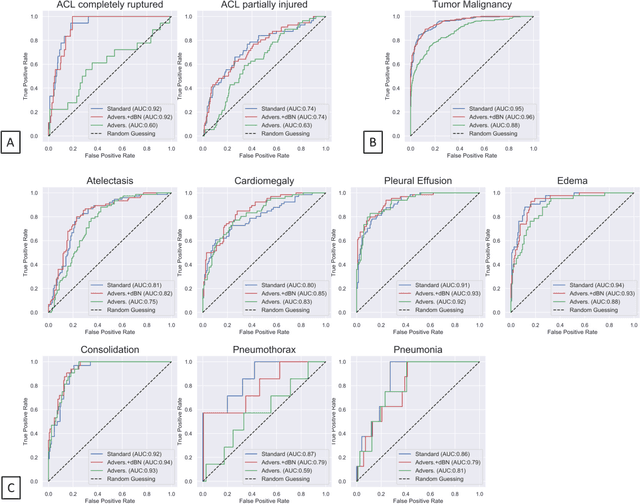

Abstract:Unmasking the decision-making process of machine learning models is essential for implementing diagnostic support systems in clinical practice. Here, we demonstrate that adversarially trained models can significantly enhance the usability of pathology detection as compared to their standard counterparts. We let six experienced radiologists rate the interpretability of saliency maps in datasets of X-rays, computed tomography, and magnetic resonance imaging scans. Significant improvements were found for our adversarial models, which could be further improved by the application of dual batch normalization. Contrary to previous research on adversarially trained models, we found that the accuracy of such models was equal to standard models when sufficiently large datasets and dual batch norm training were used. To ensure transferability, we additionally validated our results on an external test set of 22,433 X-rays. These findings elucidate that different paths for adversarial and real images are needed during training to achieve state of the art results with superior clinical interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge