Peter Isfort
Mind the Gap: Federated Learning Broadens Domain Generalization in Diagnostic AI Models
Oct 01, 2023Abstract:Developing robust artificial intelligence (AI) models that generalize well to unseen datasets is challenging and usually requires large and variable datasets, preferably from multiple institutions. In federated learning (FL), a model is trained collaboratively at numerous sites that hold local datasets without exchanging them. So far, the impact of training strategy, i.e., local versus collaborative, on the diagnostic on-domain and off-domain performance of AI models interpreting chest radiographs has not been assessed. Consequently, using 610,000 chest radiographs from five institutions across the globe, we assessed diagnostic performance as a function of training strategy (i.e., local vs. collaborative), network architecture (i.e., convolutional vs. transformer-based), generalization performance (i.e., on-domain vs. off-domain), imaging finding (i.e., cardiomegaly, pleural effusion, pneumonia, atelectasis, consolidation, pneumothorax, and no abnormality), dataset size (i.e., from n=18,000 to 213,921 radiographs), and dataset diversity. Large datasets not only showed minimal performance gains with FL but, in some instances, even exhibited decreases. In contrast, smaller datasets revealed marked improvements. Thus, on-domain performance was mainly driven by training data size. However, off-domain performance leaned more on training diversity. When trained collaboratively across diverse external institutions, AI models consistently surpassed models trained locally for off-domain tasks, emphasizing FL's potential in leveraging data diversity. In conclusion, FL can bolster diagnostic privacy, reproducibility, and off-domain reliability of AI models and, potentially, optimize healthcare outcomes.
Preserving privacy in domain transfer of medical AI models comes at no performance costs: The integral role of differential privacy
Jun 10, 2023Abstract:Developing robust and effective artificial intelligence (AI) models in medicine requires access to large amounts of patient data. The use of AI models solely trained on large multi-institutional datasets can help with this, yet the imperative to ensure data privacy remains, particularly as membership inference risks breaching patient confidentiality. As a proposed remedy, we advocate for the integration of differential privacy (DP). We specifically investigate the performance of models trained with DP as compared to models trained without DP on data from institutions that the model had not seen during its training (i.e., external validation) - the situation that is reflective of the clinical use of AI models. By leveraging more than 590,000 chest radiographs from five institutions, we evaluated the efficacy of DP-enhanced domain transfer (DP-DT) in diagnosing cardiomegaly, pleural effusion, pneumonia, atelectasis, and in identifying healthy subjects. We juxtaposed DP-DT with non-DP-DT and examined diagnostic accuracy and demographic fairness using the area under the receiver operating characteristic curve (AUC) as the main metric, as well as accuracy, sensitivity, and specificity. Our results show that DP-DT, even with exceptionally high privacy levels (epsilon around 1), performs comparably to non-DP-DT (P>0.119 across all domains). Furthermore, DP-DT led to marginal AUC differences - less than 1% - for nearly all subgroups, relative to non-DP-DT. Despite consistent evidence suggesting that DP models induce significant performance degradation for on-domain applications, we show that off-domain performance is almost not affected. Therefore, we ardently advocate for the adoption of DP in training diagnostic medical AI models, given its minimal impact on performance.
Collaborative Training of Medical Artificial Intelligence Models with non-uniform Labels
Nov 24, 2022Abstract:Artificial intelligence (AI) methods are revolutionizing medical image analysis. However, robust AI models require large multi-site datasets for training. While multiple stakeholders have provided publicly available datasets, the ways in which these data are labeled differ widely. For example, one dataset of chest radiographs might contain labels denoting the presence of metastases in the lung, while another dataset of chest radiograph might focus on the presence of pneumonia. With conventional approaches, these data cannot be used together to train a single AI model. We propose a new framework that we call flexible federated learning (FFL) for collaborative training on such data. Using publicly available data of 695,000 chest radiographs from five institutions - each with differing labels - we demonstrate that large and heterogeneously labeled datasets can be used to train one big AI model with this framework. We find that models trained with FFL are superior to models that are trained on matching annotations only. This may pave the way for training of truly large-scale AI models that make efficient use of all existing data.
Predicting Osteoarthritis Progression in Radiographs via Unsupervised Representation Learning
Nov 22, 2021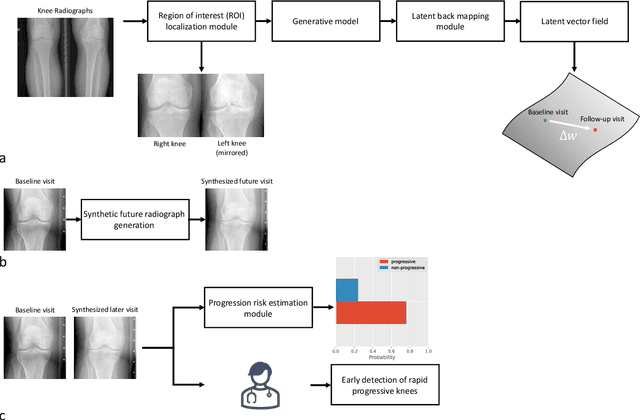
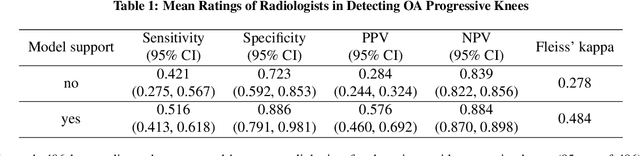
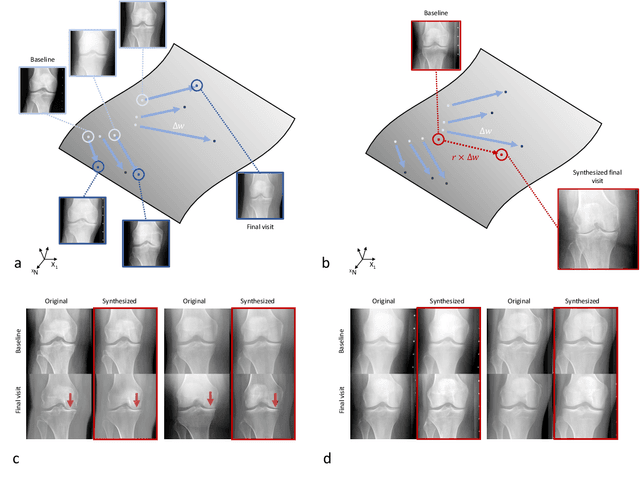
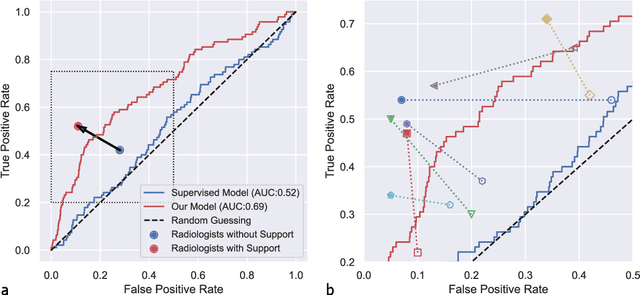
Abstract:Osteoarthritis (OA) is the most common joint disorder affecting substantial proportions of the global population, primarily the elderly. Despite its individual and socioeconomic burden, the onset and progression of OA can still not be reliably predicted. Aiming to fill this diagnostic gap, we introduce an unsupervised learning scheme based on generative models to predict the future development of OA based on knee joint radiographs. Using longitudinal data from osteoarthritis studies, we explore the latent temporal trajectory to predict a patient's future radiographs up to the eight-year follow-up visit. Our model predicts the risk of progression towards OA and surpasses its supervised counterpart whose input was provided by seven experienced radiologists. With the support of the model, sensitivity, specificity, positive predictive value, and negative predictive value increased significantly from 42.1% to 51.6%, from 72.3% to 88.6%, from 28.4% to 57.6%, and from 83.9% to 88.4%, respectively, while without such support, radiologists performed only slightly better than random guessing. Our predictive model improves predictions on OA onset and progression, despite requiring no human annotation in the training phase.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge