Sven Nebelung
The role of self-supervised pretraining in differentially private medical image analysis
Jan 27, 2026Abstract:Differential privacy (DP) provides formal protection for sensitive data but typically incurs substantial losses in diagnostic performance. Model initialization has emerged as a critical factor in mitigating this degradation, yet the role of modern self-supervised learning under full-model DP remains poorly understood. Here, we present a large-scale evaluation of initialization strategies for differentially private medical image analysis, using chest radiograph classification as a representative benchmark with more than 800,000 images. Using state-of-the-art ConvNeXt models trained with DP-SGD across realistic privacy regimes, we compare non-domain-specific supervised ImageNet initialization, non-domain-specific self-supervised DINOv3 initialization, and domain-specific supervised pretraining on MIMIC-CXR, the largest publicly available chest radiograph dataset. Evaluations are conducted across five external datasets spanning diverse institutions and acquisition settings. We show that DINOv3 initialization consistently improves diagnostic utility relative to ImageNet initialization under DP, but remains inferior to domain-specific supervised pretraining, which achieves performance closest to non-private baselines. We further demonstrate that initialization choice strongly influences demographic fairness, cross-dataset generalization, and robustness to data scale and model capacity under privacy constraints. The results establish initialization strategy as a central determinant of utility, fairness, and generalization in differentially private medical imaging.
Resolution scaling governs DINOv3 transfer performance in chest radiograph classification
Oct 08, 2025Abstract:Self-supervised learning (SSL) has advanced visual representation learning, but its value in chest radiography, a high-volume imaging modality with fine-grained findings, remains unclear. Meta's DINOv3 extends earlier SSL models through Gram-anchored self-distillation. Whether these design choices improve transfer learning for chest radiography has not been systematically tested. We benchmarked DINOv3 against DINOv2 and ImageNet initialization across seven datasets (n>814,000). Two representative backbones were evaluated: ViT-B/16 and ConvNeXt-B. Images were analyzed at 224x224, 512x512, and 1024x1024 pixels. We additionally assessed frozen features from a 7B model. The primary outcome was mean AUROC across labels. At 224x224, DINOv3 and DINOv2 achieved comparable performance on adult datasets. Increasing resolution to 512x512 yielded consistent improvements for DINOv3 over both DINOv2 and ImageNet. In contrast, results in pediatric cohort showed no differences across initializations. Across all settings, ConvNeXt-B outperformed ViT-B/16. Models using frozen DINOv3-7B features underperformed relative to fully finetuned 86-89M-parameter backbones, highlighting the importance of domain adaptation. Scaling to 1024x1024 did not further improve accuracy. Resolution-related gains were most evident for boundary-dependent and small focal abnormalities. In chest radiography, higher input resolution is critical for leveraging the benefits of modern self-supervised models. 512x512 pixels represent a practical upper limit where DINOv3-initialized ConvNeXt-B networks provide the strongest performance, while larger inputs offer minimal return on cost. Clinically, these findings support use of finetuned, mid-sized backbones at 512x512 for chest radiograph interpretation, with the greatest gains expected in detecting subtle or boundary-centered lesions relevant to emergency and critical care settings.
MedicalPatchNet: A Patch-Based Self-Explainable AI Architecture for Chest X-ray Classification
Sep 09, 2025Abstract:Deep neural networks excel in radiological image classification but frequently suffer from poor interpretability, limiting clinical acceptance. We present MedicalPatchNet, an inherently self-explainable architecture for chest X-ray classification that transparently attributes decisions to distinct image regions. MedicalPatchNet splits images into non-overlapping patches, independently classifies each patch, and aggregates predictions, enabling intuitive visualization of each patch's diagnostic contribution without post-hoc techniques. Trained on the CheXpert dataset (223,414 images), MedicalPatchNet matches the classification performance (AUROC 0.907 vs. 0.908) of EfficientNet-B0, while substantially improving interpretability: MedicalPatchNet demonstrates substantially improved interpretability with higher pathology localization accuracy (mean hit-rate 0.485 vs. 0.376 with Grad-CAM) on the CheXlocalize dataset. By providing explicit, reliable explanations accessible even to non-AI experts, MedicalPatchNet mitigates risks associated with shortcut learning, thus improving clinical trust. Our model is publicly available with reproducible training and inference scripts and contributes to safer, explainable AI-assisted diagnostics across medical imaging domains. We make the code publicly available: https://github.com/TruhnLab/MedicalPatchNet
Medical Slice Transformer: Improved Diagnosis and Explainability on 3D Medical Images with DINOv2
Nov 24, 2024



Abstract:MRI and CT are essential clinical cross-sectional imaging techniques for diagnosing complex conditions. However, large 3D datasets with annotations for deep learning are scarce. While methods like DINOv2 are encouraging for 2D image analysis, these methods have not been applied to 3D medical images. Furthermore, deep learning models often lack explainability due to their "black-box" nature. This study aims to extend 2D self-supervised models, specifically DINOv2, to 3D medical imaging while evaluating their potential for explainable outcomes. We introduce the Medical Slice Transformer (MST) framework to adapt 2D self-supervised models for 3D medical image analysis. MST combines a Transformer architecture with a 2D feature extractor, i.e., DINOv2. We evaluate its diagnostic performance against a 3D convolutional neural network (3D ResNet) across three clinical datasets: breast MRI (651 patients), chest CT (722 patients), and knee MRI (1199 patients). Both methods were tested for diagnosing breast cancer, predicting lung nodule dignity, and detecting meniscus tears. Diagnostic performance was assessed by calculating the Area Under the Receiver Operating Characteristic Curve (AUC). Explainability was evaluated through a radiologist's qualitative comparison of saliency maps based on slice and lesion correctness. P-values were calculated using Delong's test. MST achieved higher AUC values compared to ResNet across all three datasets: breast (0.94$\pm$0.01 vs. 0.91$\pm$0.02, P=0.02), chest (0.95$\pm$0.01 vs. 0.92$\pm$0.02, P=0.13), and knee (0.85$\pm$0.04 vs. 0.69$\pm$0.05, P=0.001). Saliency maps were consistently more precise and anatomically correct for MST than for ResNet. Self-supervised 2D models like DINOv2 can be effectively adapted for 3D medical imaging using MST, offering enhanced diagnostic accuracy and explainability compared to convolutional neural networks.
RadioRAG: Factual Large Language Models for Enhanced Diagnostics in Radiology Using Dynamic Retrieval Augmented Generation
Jul 22, 2024



Abstract:Large language models (LLMs) have advanced the field of artificial intelligence (AI) in medicine. However LLMs often generate outdated or inaccurate information based on static training datasets. Retrieval augmented generation (RAG) mitigates this by integrating outside data sources. While previous RAG systems used pre-assembled, fixed databases with limited flexibility, we have developed Radiology RAG (RadioRAG) as an end-to-end framework that retrieves data from authoritative radiologic online sources in real-time. RadioRAG is evaluated using a dedicated radiologic question-and-answer dataset (RadioQA). We evaluate the diagnostic accuracy of various LLMs when answering radiology-specific questions with and without access to additional online information via RAG. Using 80 questions from RSNA Case Collection across radiologic subspecialties and 24 additional expert-curated questions, for which the correct gold-standard answers were available, LLMs (GPT-3.5-turbo, GPT-4, Mistral-7B, Mixtral-8x7B, and Llama3 [8B and 70B]) were prompted with and without RadioRAG. RadioRAG retrieved context-specific information from www.radiopaedia.org in real-time and incorporated them into its reply. RadioRAG consistently improved diagnostic accuracy across all LLMs, with relative improvements ranging from 2% to 54%. It matched or exceeded question answering without RAG across radiologic subspecialties, particularly in breast imaging and emergency radiology. However, degree of improvement varied among models; GPT-3.5-turbo and Mixtral-8x7B-instruct-v0.1 saw notable gains, while Mistral-7B-instruct-v0.2 showed no improvement, highlighting variability in its effectiveness. LLMs benefit when provided access to domain-specific data beyond their training data. For radiology, RadioRAG establishes a robust framework that substantially improves diagnostic accuracy and factuality in radiological question answering.
On Instabilities of Unsupervised Denoising Diffusion Models in Magnetic Resonance Imaging Reconstruction
Jun 23, 2024Abstract:Denoising diffusion models offer a promising approach to accelerating magnetic resonance imaging (MRI) and producing diagnostic-level images in an unsupervised manner. However, our study demonstrates that even tiny worst-case potential perturbations transferred from a surrogate model can cause these models to generate fake tissue structures that may mislead clinicians. The transferability of such worst-case perturbations indicates that the robustness of image reconstruction may be compromised due to MR system imperfections or other sources of noise. Moreover, at larger perturbation strengths, diffusion models exhibit Gaussian noise-like artifacts that are distinct from those observed in supervised models and are more challenging to detect. Our results highlight the vulnerability of current state-of-the-art diffusion-based reconstruction models to possible worst-case perturbations and underscore the need for further research to improve their robustness and reliability in clinical settings.
Compute-Efficient Medical Image Classification with Softmax-Free Transformers and Sequence Normalization
Jun 03, 2024Abstract:The Transformer model has been pivotal in advancing fields such as natural language processing, speech recognition, and computer vision. However, a critical limitation of this model is its quadratic computational and memory complexity relative to the sequence length, which constrains its application to longer sequences. This is especially crucial in medical imaging where high-resolution images can reach gigapixel scale. Efforts to address this issue have predominantely focused on complex techniques, such as decomposing the softmax operation integral to the Transformer's architecture. This paper addresses this quadratic computational complexity of Transformer models and introduces a remarkably simple and effective method that circumvents this issue by eliminating the softmax function from the attention mechanism and adopting a sequence normalization technique for the key, query, and value tokens. Coupled with a reordering of matrix multiplications this approach reduces the memory- and compute complexity to a linear scale. We evaluate this approach across various medical imaging datasets comprising fundoscopic, dermascopic, radiologic and histologic imaging data. Our findings highlight that these models exhibit a comparable performance to traditional transformer models, while efficiently handling longer sequences.
An Ordinal Regression Framework for a Deep Learning Based Severity Assessment for Chest Radiographs
Feb 08, 2024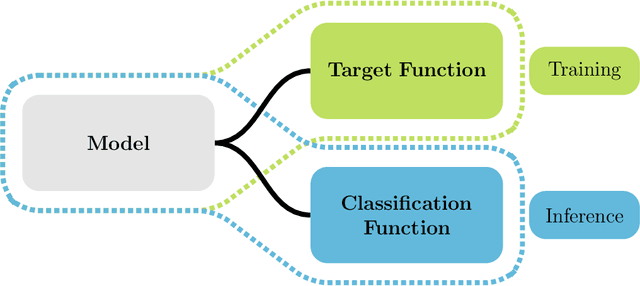
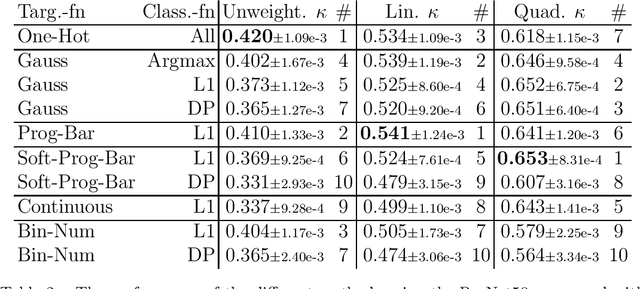
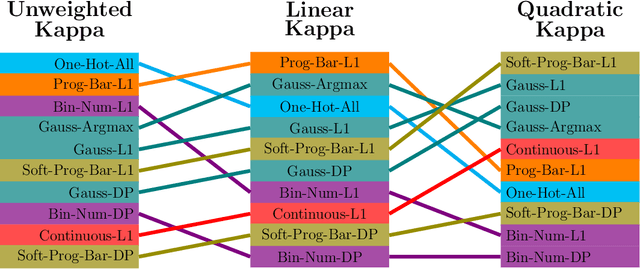
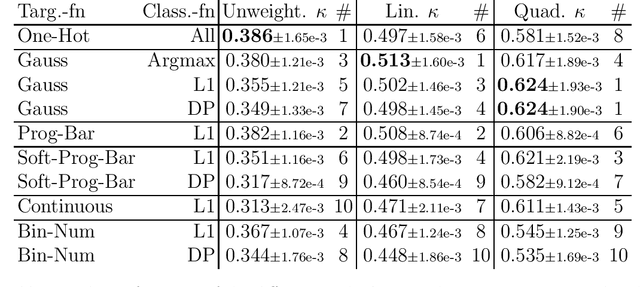
Abstract:This study investigates the application of ordinal regression methods for categorizing disease severity in chest radiographs. We propose a framework that divides the ordinal regression problem into three parts: a model, a target function, and a classification function. Different encoding methods, including one-hot, Gaussian, progress-bar, and our soft-progress-bar, are applied using ResNet50 and ViT-B-16 deep learning models. We show that the choice of encoding has a strong impact on performance and that the best encoding depends on the chosen weighting of Cohen's kappa and also on the model architecture used. We make our code publicly available on GitHub.
Time-efficient combined morphologic and quantitative joint MRI based on clinical image contrasts -- An exploratory in-situ study of standardized cartilage defects
Nov 14, 2023Abstract:OBJECTIVES: Quantitative MRI techniques such as T2 and T1$\rho$ mapping are beneficial in evaluating cartilage and meniscus. We aimed to evaluate the MIXTURE (Multi-Interleaved X-prepared Turbo-Spin Echo with IntUitive RElaxometry) sequences that provide morphologic images with clinical turbo spin-echo (TSE) contrasts and additional parameter maps versus reference TSE sequences in an in-situ model of human cartilage defects. MATERIALS AND METHODS: Prospectively, standardized cartilage defects of 8mm, 5mm, and 3mm diameter were created in the lateral femora of 10 human cadaveric knee specimens (81$\pm$10 years, nine male/one female). Using a clinical 3T MRI scanner and knee coil, MIXTURE sequences combining (i) proton-density weighted fat-saturated (PD-w FS) images and T2 maps and (ii) T1-weighted images and T1$\rho$ maps were acquired before and after defect creation, alongside the corresponding 2D TSE and 3D TSE reference sequences. Defect delineability, bone texture, and cartilage relaxation times were quantified. Inter-sequence comparisons were made using appropriate parametric and non-parametric tests. RESULTS: Overall, defect delineability and texture features were not significantly different between the MIXTURE and reference sequences. After defect creation, relaxation times increased significantly in the central femur (for T2) and all regions combined (for T1$\rho$). CONCLUSION: MIXTURE sequences permit time-efficient simultaneous morphologic and quantitative joint assessment based on clinical image contrasts. While providing T2 or T1$\rho$ maps in clinically feasible scan time, morphologic image features, i.e., cartilage defect delineability and bone texture, were comparable between MIXTURE and corresponding reference sequences.
Mind the Gap: Federated Learning Broadens Domain Generalization in Diagnostic AI Models
Oct 01, 2023Abstract:Developing robust artificial intelligence (AI) models that generalize well to unseen datasets is challenging and usually requires large and variable datasets, preferably from multiple institutions. In federated learning (FL), a model is trained collaboratively at numerous sites that hold local datasets without exchanging them. So far, the impact of training strategy, i.e., local versus collaborative, on the diagnostic on-domain and off-domain performance of AI models interpreting chest radiographs has not been assessed. Consequently, using 610,000 chest radiographs from five institutions across the globe, we assessed diagnostic performance as a function of training strategy (i.e., local vs. collaborative), network architecture (i.e., convolutional vs. transformer-based), generalization performance (i.e., on-domain vs. off-domain), imaging finding (i.e., cardiomegaly, pleural effusion, pneumonia, atelectasis, consolidation, pneumothorax, and no abnormality), dataset size (i.e., from n=18,000 to 213,921 radiographs), and dataset diversity. Large datasets not only showed minimal performance gains with FL but, in some instances, even exhibited decreases. In contrast, smaller datasets revealed marked improvements. Thus, on-domain performance was mainly driven by training data size. However, off-domain performance leaned more on training diversity. When trained collaboratively across diverse external institutions, AI models consistently surpassed models trained locally for off-domain tasks, emphasizing FL's potential in leveraging data diversity. In conclusion, FL can bolster diagnostic privacy, reproducibility, and off-domain reliability of AI models and, potentially, optimize healthcare outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge