Marc Huppertz
Time-efficient combined morphologic and quantitative joint MRI based on clinical image contrasts -- An exploratory in-situ study of standardized cartilage defects
Nov 14, 2023Abstract:OBJECTIVES: Quantitative MRI techniques such as T2 and T1$\rho$ mapping are beneficial in evaluating cartilage and meniscus. We aimed to evaluate the MIXTURE (Multi-Interleaved X-prepared Turbo-Spin Echo with IntUitive RElaxometry) sequences that provide morphologic images with clinical turbo spin-echo (TSE) contrasts and additional parameter maps versus reference TSE sequences in an in-situ model of human cartilage defects. MATERIALS AND METHODS: Prospectively, standardized cartilage defects of 8mm, 5mm, and 3mm diameter were created in the lateral femora of 10 human cadaveric knee specimens (81$\pm$10 years, nine male/one female). Using a clinical 3T MRI scanner and knee coil, MIXTURE sequences combining (i) proton-density weighted fat-saturated (PD-w FS) images and T2 maps and (ii) T1-weighted images and T1$\rho$ maps were acquired before and after defect creation, alongside the corresponding 2D TSE and 3D TSE reference sequences. Defect delineability, bone texture, and cartilage relaxation times were quantified. Inter-sequence comparisons were made using appropriate parametric and non-parametric tests. RESULTS: Overall, defect delineability and texture features were not significantly different between the MIXTURE and reference sequences. After defect creation, relaxation times increased significantly in the central femur (for T2) and all regions combined (for T1$\rho$). CONCLUSION: MIXTURE sequences permit time-efficient simultaneous morphologic and quantitative joint assessment based on clinical image contrasts. While providing T2 or T1$\rho$ maps in clinically feasible scan time, morphologic image features, i.e., cartilage defect delineability and bone texture, were comparable between MIXTURE and corresponding reference sequences.
Reconstruction of Patient-Specific Confounders in AI-based Radiologic Image Interpretation using Generative Pretraining
Sep 29, 2023
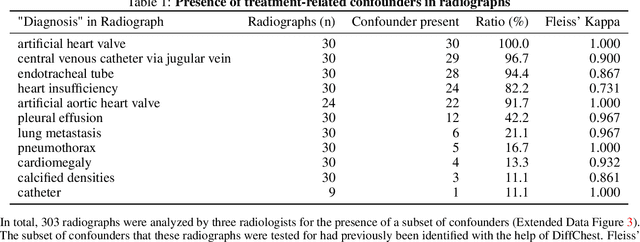
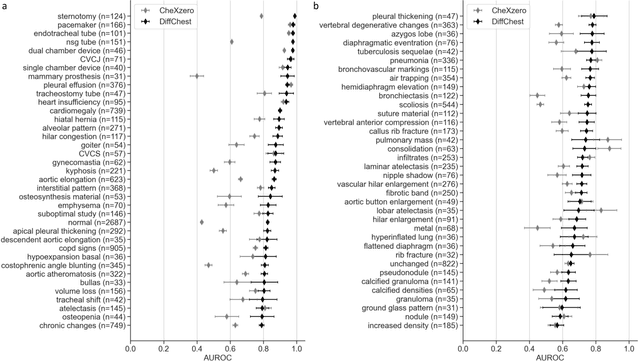
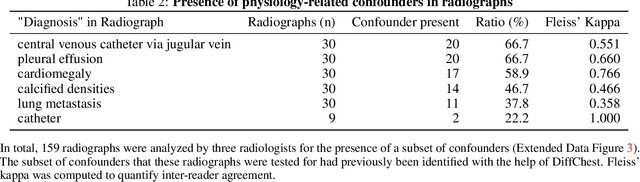
Abstract:Detecting misleading patterns in automated diagnostic assistance systems, such as those powered by Artificial Intelligence, is critical to ensuring their reliability, particularly in healthcare. Current techniques for evaluating deep learning models cannot visualize confounding factors at a diagnostic level. Here, we propose a self-conditioned diffusion model termed DiffChest and train it on a dataset of 515,704 chest radiographs from 194,956 patients from multiple healthcare centers in the United States and Europe. DiffChest explains classifications on a patient-specific level and visualizes the confounding factors that may mislead the model. We found high inter-reader agreement when evaluating DiffChest's capability to identify treatment-related confounders, with Fleiss' Kappa values of 0.8 or higher across most imaging findings. Confounders were accurately captured with 11.1% to 100% prevalence rates. Furthermore, our pretraining process optimized the model to capture the most relevant information from the input radiographs. DiffChest achieved excellent diagnostic accuracy when diagnosing 11 chest conditions, such as pleural effusion and cardiac insufficiency, and at least sufficient diagnostic accuracy for the remaining conditions. Our findings highlight the potential of pretraining based on diffusion models in medical image classification, specifically in providing insights into confounding factors and model robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge