Volkmar Schulz
Diffusion-based Sinogram Interpolation for Limited Angle PET
Nov 12, 2025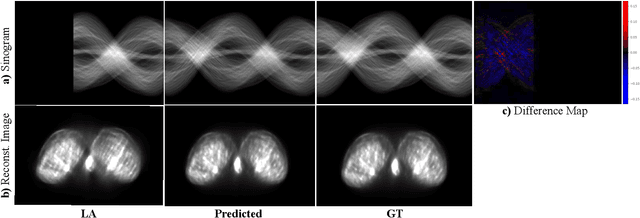
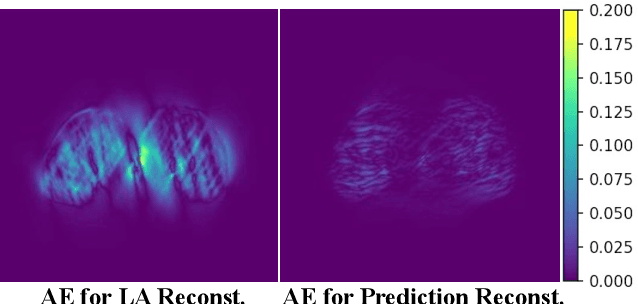
Abstract:Accurate PET imaging increasingly requires methods that support unconstrained detector layouts from walk-through designs to long-axial rings where gaps and open sides lead to severely undersampled sinograms. Instead of constraining the hardware to form complete cylinders, we propose treating the missing lines-of-responses as a learnable prior. Data-driven approaches, particularly generative models, offer a promising pathway to recover this missing information. In this work, we explore the use of conditional diffusion models to interpolate sparsely sampled sinograms, paving the way for novel, cost-efficient, and patient-friendly PET geometries in real clinical settings.
Towards Robust and Generalizable Gerchberg Saxton based Physics Inspired Neural Networks for Computer Generated Holography: A Sensitivity Analysis Framework
Apr 30, 2025Abstract:Computer-generated holography (CGH) enables applications in holographic augmented reality (AR), 3D displays, systems neuroscience, and optical trapping. The fundamental challenge in CGH is solving the inverse problem of phase retrieval from intensity measurements. Physics-inspired neural networks (PINNs), especially Gerchberg-Saxton-based PINNs (GS-PINNs), have advanced phase retrieval capabilities. However, their performance strongly depends on forward models (FMs) and their hyperparameters (FMHs), limiting generalization, complicating benchmarking, and hindering hardware optimization. We present a systematic sensitivity analysis framework based on Saltelli's extension of Sobol's method to quantify FMH impacts on GS-PINN performance. Our analysis demonstrates that SLM pixel-resolution is the primary factor affecting neural network sensitivity, followed by pixel-pitch, propagation distance, and wavelength. Free space propagation forward models demonstrate superior neural network performance compared to Fourier holography, providing enhanced parameterization and generalization. We introduce a composite evaluation metric combining performance consistency, generalization capability, and hyperparameter perturbation resilience, establishing a unified benchmarking standard across CGH configurations. Our research connects physics-inspired deep learning theory with practical CGH implementations through concrete guidelines for forward model selection, neural network architecture, and performance evaluation. Our contributions advance the development of robust, interpretable, and generalizable neural networks for diverse holographic applications, supporting evidence-based decisions in CGH research and implementation.
Rethinking Timing Residuals: Advancing PET Detectors with Explicit TOF Corrections
Feb 11, 2025Abstract:PET is a functional imaging method that visualizes metabolic processes. TOF information can be derived from coincident detector signals and incorporated into image reconstruction to enhance the SNR. PET detectors are typically assessed by their CTR, but timing performance is degraded by various factors. Research on timing calibration seeks to mitigate these degradations and restore accurate timing information. While many calibration methods use analytical approaches, machine learning techniques have recently gained attention due to their flexibility. We developed a residual physics-based calibration approach that combines prior domain knowledge with the power of machine learning models. This approach begins with an initial analytical calibration addressing first-order skews. The remaining deviations, regarded as residual effects, are used to train machine learning models to eliminate higher-order skews. The key advantage is that the experimenter guides the learning process through the definition of timing residuals. In earlier studies, we developed models that directly predicted the expected time difference, which offered corrections only implicitly (implicit correction models). In this study, we introduce a new definition for timing residuals, enabling us to train models that directly predict correction values (explicit correction models). The explicit correction approach significantly simplifies data acquisition, improves linearity, and enhances timing performance from $371 \pm 6$ ps to $281 \pm 5$ ps for coincidences from 430 keV to 590 keV. Additionally, the new definition reduces model size, making it suitable for high-throughput applications like PET scanners. Experiments were conducted using two detector stacks composed of $4 \times 4$ LYSO:Ce,Ca crystals ($3.8\times 3.8\times 20$ mm$^{3}$) coupled to $4 \times 4$ Broadcom NUV-MT SiPMs and digitized with the TOFPET2 ASIC.
Overcoming Data Scarcity in Biomedical Imaging with a Foundational Multi-Task Model
Nov 16, 2023Abstract:Foundational models, pretrained on a large scale, have demonstrated substantial success across non-medical domains. However, training these models typically requires large, comprehensive datasets, which contrasts with the smaller and more heterogeneous datasets common in biomedical imaging. Here, we propose a multi-task learning strategy that decouples the number of training tasks from memory requirements. We trained a Universal bioMedical PreTrained model (UMedPT) on a multi-task database including tomographic, microscopic, and X-ray images, with various labelling strategies such as classification, segmentation, and object detection. The UMedPT foundational model outperformed ImageNet pretraining and the previous state-of-the-art models. For tasks related to the pretraining database, it maintained its performance with only 1% of the original training data and without fine-tuning. For out-of-domain tasks it required not more than 50% of the original training data. In an external independent validation imaging features extracted using UMedPT proved to be a new standard for cross-center transferability.
Improving the Timing Resolution of Positron Emission Tomography Detectors using Boosted Learning -- A Residual Physics Approach
Feb 03, 2023



Abstract:Artificial intelligence is finding its way into medical imaging, usually focusing on image reconstruction or enhancing analytical reconstructed images. However, optimizations along the complete processing chain, from detecting signals to computing data, enable significant improvements. Thus, we present an approach toward detector optimization using boosted learning by exploiting the concept of residual physics. In our work, we improve the coincidence time resolution (CTR) of positron emission tomography (PET) detectors. PET enables imaging of metabolic processes by detecting {\gamma}-photons with scintillation detectors. Current research exploits light-sharing detectors, where the scintillation light is distributed over and digitized by an array of readout channels. While these detectors demonstrate excellent performance parameters, e.g., regarding spatial resolution, extracting precise timing information for time-of-flight (TOF) becomes more challenging due to deteriorating effects called time skews. Conventional correction methods mainly rely on analytical formulations, theoretically capable of covering all time skew effects, e.g., caused by signal runtimes or physical effects. However, additional effects are involved for light-sharing detectors, so finding suitable analytical formulations can become arbitrarily complicated. The residual physics-based strategy uses gradient tree boosting (GTB) and a physics-informed data generation mimicking an actual imaging process by shifting a radiation source. We used clinically relevant detectors with a height of 19 mm, coupled to digital photosensor arrays. All trained models improved the CTR significantly. Using the best model, we achieved CTRs down to 198 ps (185 ps) for energies ranging from 300 keV to 700 keV (450 keV to 550 keV).
Predicting Osteoarthritis Progression in Radiographs via Unsupervised Representation Learning
Nov 22, 2021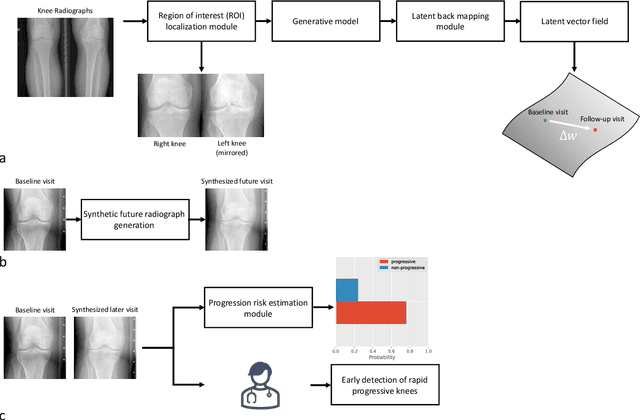
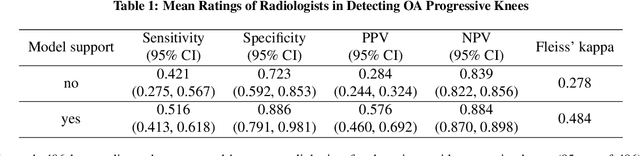
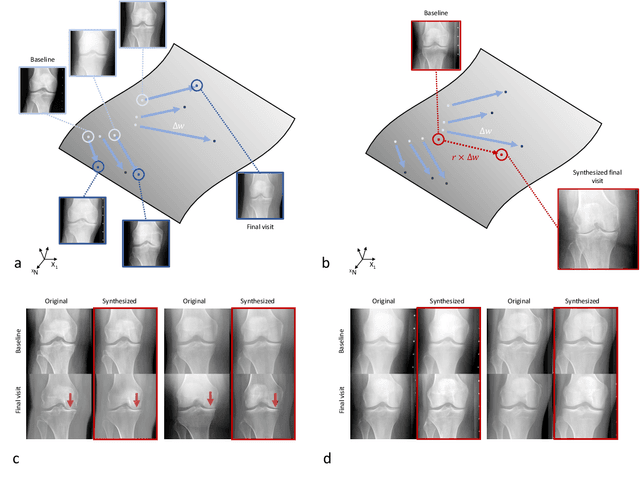
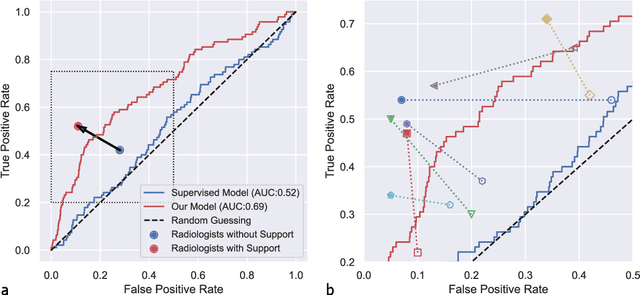
Abstract:Osteoarthritis (OA) is the most common joint disorder affecting substantial proportions of the global population, primarily the elderly. Despite its individual and socioeconomic burden, the onset and progression of OA can still not be reliably predicted. Aiming to fill this diagnostic gap, we introduce an unsupervised learning scheme based on generative models to predict the future development of OA based on knee joint radiographs. Using longitudinal data from osteoarthritis studies, we explore the latent temporal trajectory to predict a patient's future radiographs up to the eight-year follow-up visit. Our model predicts the risk of progression towards OA and surpasses its supervised counterpart whose input was provided by seven experienced radiologists. With the support of the model, sensitivity, specificity, positive predictive value, and negative predictive value increased significantly from 42.1% to 51.6%, from 72.3% to 88.6%, from 28.4% to 57.6%, and from 83.9% to 88.4%, respectively, while without such support, radiologists performed only slightly better than random guessing. Our predictive model improves predictions on OA onset and progression, despite requiring no human annotation in the training phase.
Advancing diagnostic performance and clinical usability of neural networks via adversarial training and dual batch normalization
Nov 25, 2020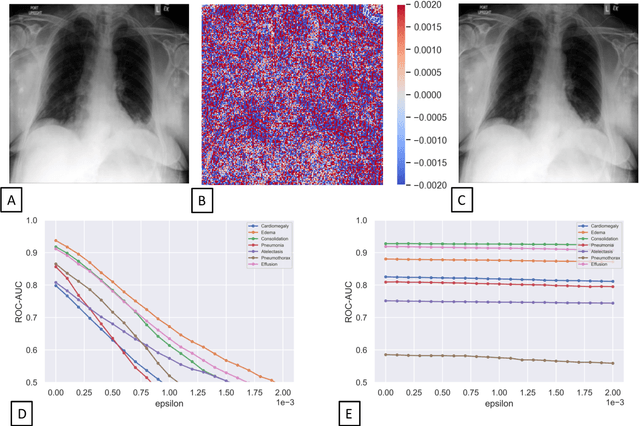
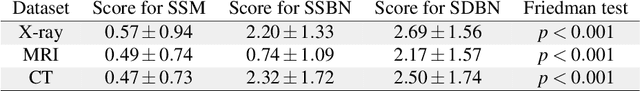
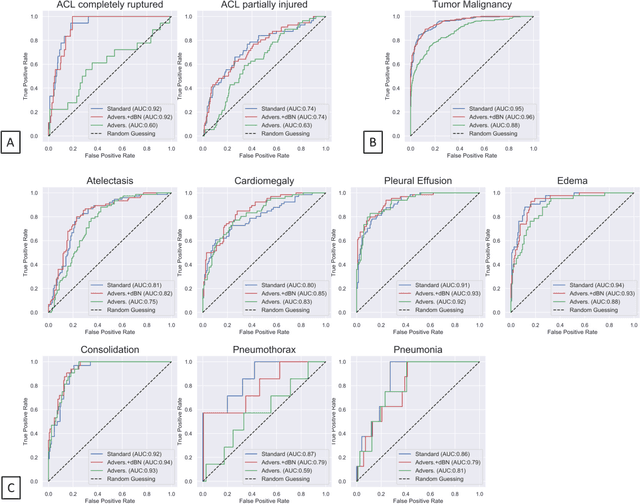

Abstract:Unmasking the decision-making process of machine learning models is essential for implementing diagnostic support systems in clinical practice. Here, we demonstrate that adversarially trained models can significantly enhance the usability of pathology detection as compared to their standard counterparts. We let six experienced radiologists rate the interpretability of saliency maps in datasets of X-rays, computed tomography, and magnetic resonance imaging scans. Significant improvements were found for our adversarial models, which could be further improved by the application of dual batch normalization. Contrary to previous research on adversarially trained models, we found that the accuracy of such models was equal to standard models when sufficiently large datasets and dual batch norm training were used. To ensure transferability, we additionally validated our results on an external test set of 22,433 X-rays. These findings elucidate that different paths for adversarial and real images are needed during training to achieve state of the art results with superior clinical interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge