Zijing Dong
Latent Signal Models: Learning Compact Representations of Signal Evolution for Improved Time-Resolved, Multi-contrast MRI
Aug 27, 2022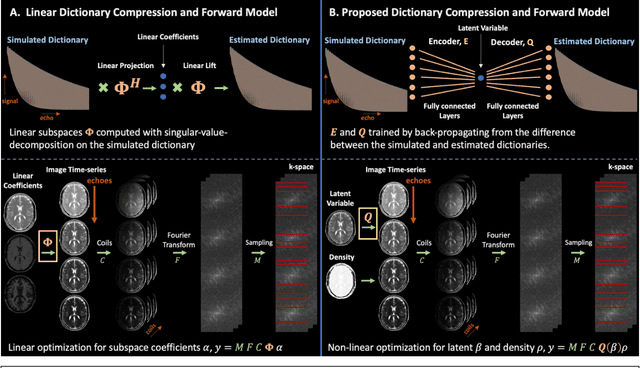
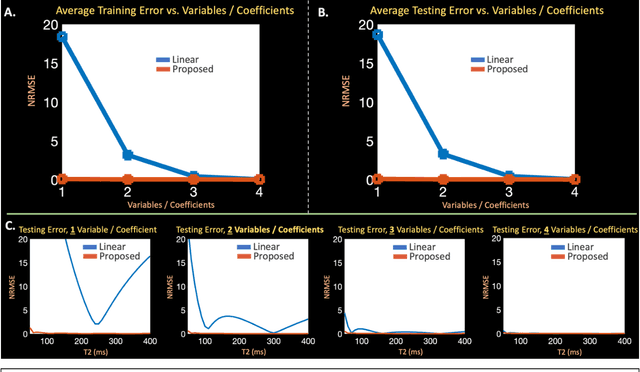
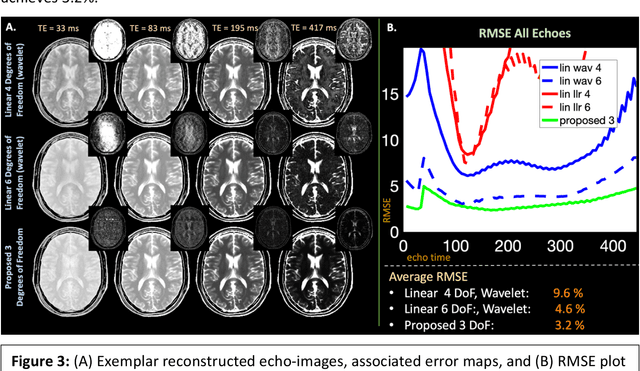
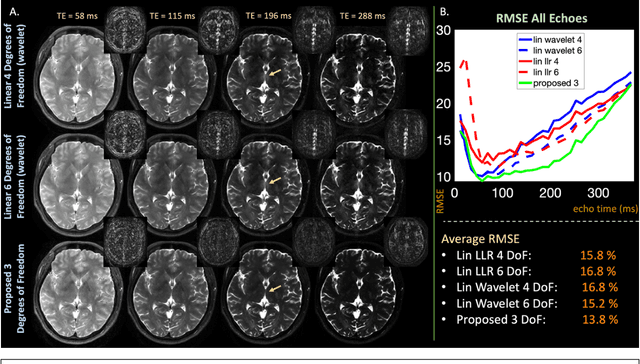
Abstract:Purpose: Training auto-encoders on simulated signal evolution and inserting the decoder into the forward model improves reconstructions through more compact, Bloch-equation-based representations of signal in comparison to linear subspaces. Methods: Building on model-based nonlinear and linear subspace techniques that enable reconstruction of signal dynamics, we train auto-encoders on dictionaries of simulated signal evolution to learn more compact, non-linear, latent representations. The proposed Latent Signal Model framework inserts the decoder portion of the auto-encoder into the forward model and directly reconstructs the latent representation. Latent Signal Models essentially serve as a proxy for fast and feasible differentiation through the Bloch-equations used to simulate signal. This work performs experiments in the context of T2-shuffling, gradient echo EPTI, and MPRAGE-shuffling. We compare how efficiently auto-encoders represent signal evolution in comparison to linear subspaces. Simulation and in-vivo experiments then evaluate if reducing degrees of freedom by inserting the decoder into the forward model improves reconstructions in comparison to subspace constraints. Results: An auto-encoder with one real latent variable represents FSE, EPTI, and MPRAGE signal evolution as well as linear subspaces characterized by four basis vectors. In simulated/in-vivo T2-shuffling and in-vivo EPTI experiments, the proposed framework achieves consistent quantitative NRMSE and qualitative improvement over linear approaches. From qualitative evaluation, the proposed approach yields images with reduced blurring and noise amplification in MPRAGE shuffling experiments. Conclusion: Directly solving for non-linear latent representations of signal evolution improves time-resolved MRI reconstructions through reduced degrees of freedom.
Optimized multi-axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole-brain high-isotropic-resolution quantitative imaging
Aug 12, 2021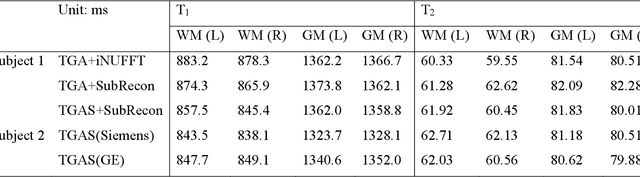
Abstract:Purpose: To improve image quality and accelerate the acquisition of 3D MRF. Methods: Building on the multi-axis spiral-projection MRF technique, a subspace reconstruction with locally low rank (LLR) constraint and a modified spiral-projection spatiotemporal encoding scheme termed tiny-golden-angle-shuffling (TGAS) were implemented for rapid whole-brain high-resolution quantitative mapping. The LLR regularization parameter and the number of subspace bases were tuned using retrospective in-vivo data and simulated examinations, respectively. B0 inhomogeneity correction using multi-frequency interpolation was incorporated into the subspace reconstruction to further improve the image quality by mitigating blurring caused by off-resonance effect. Results: The proposed MRF acquisition and reconstruction framework can produce provide high quality 1-mm isotropic whole-brain quantitative maps in a total acquisition time of 1 minute 55 seconds, with higher-quality results than ones obtained from the previous approach in 6 minutes. The comparison of quantitative results indicates that neither the subspace reconstruction nor the TGAS trajectory induce bias for T1 and T2 mapping. High quality whole-brain MRF data were also obtained at 0.66-mm isotropic resolution in 4 minutes using the proposed technique, where the increased resolution was shown to improve visualization of subtle brain structures. Conclusion: The proposed TGAS-SPI-MRF with optimized spiral-projection trajectory and subspace reconstruction can enable high-resolution quantitative mapping with faster acquisition speed.
eRAKI: Fast Robust Artificial neural networks for K-space Interpolation (RAKI) with Coil Combination and Joint Reconstruction
Jul 07, 2021Abstract:RAKI can perform database-free MRI reconstruction by training models using only auto-calibration signal (ACS) from each specific scan. As it trains a separate model for each individual coil, learning and inference with RAKI can be computationally prohibitive, particularly for large 3D datasets. In this abstract, we accelerate RAKI more than 200 times by directly learning a coil-combined target and further improve the reconstruction performance using joint reconstruction across multiple echoes together with an elliptical-CAIPI sampling approach. We further deploy these improvements in quantitative imaging and rapidly obtain T2 and T2* parameter maps from a fast EPTI scan.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge