Siddharth Srinivasan Iyer
Group Diffusion: Enhancing Image Generation by Unlocking Cross-Sample Collaboration
Dec 11, 2025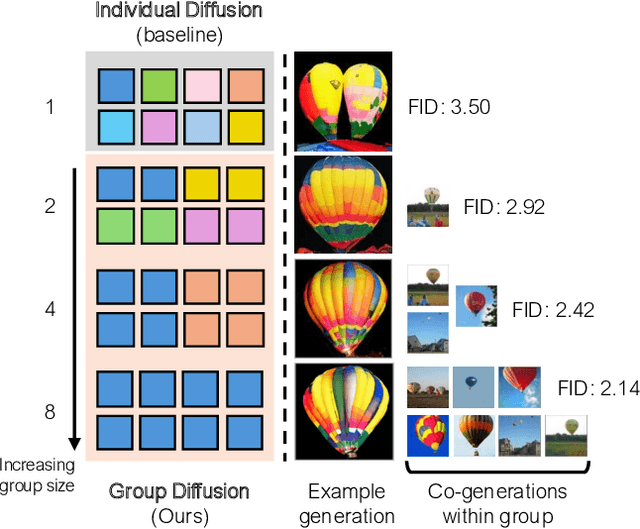
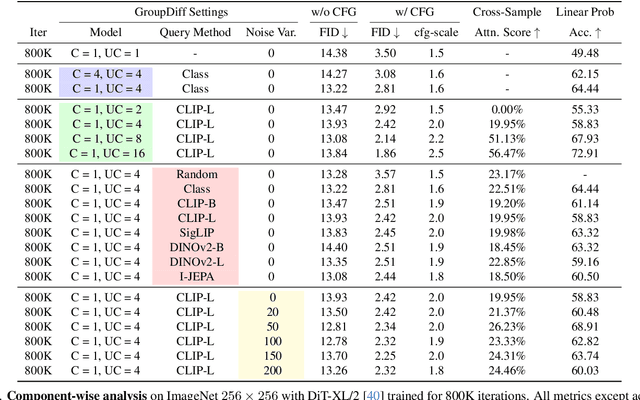

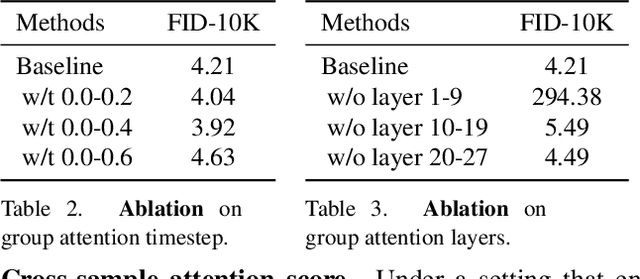
Abstract:In this work, we explore an untapped signal in diffusion model inference. While all previous methods generate images independently at inference, we instead ask if samples can be generated collaboratively. We propose Group Diffusion, unlocking the attention mechanism to be shared across images, rather than limited to just the patches within an image. This enables images to be jointly denoised at inference time, learning both intra and inter-image correspondence. We observe a clear scaling effect - larger group sizes yield stronger cross-sample attention and better generation quality. Furthermore, we introduce a qualitative measure to capture this behavior and show that its strength closely correlates with FID. Built on standard diffusion transformers, our GroupDiff achieves up to 32.2% FID improvement on ImageNet-256x256. Our work reveals cross-sample inference as an effective, previously unexplored mechanism for generative modeling.
X-Fusion: Introducing New Modality to Frozen Large Language Models
Apr 29, 2025Abstract:We propose X-Fusion, a framework that extends pretrained Large Language Models (LLMs) for multimodal tasks while preserving their language capabilities. X-Fusion employs a dual-tower design with modality-specific weights, keeping the LLM's parameters frozen while integrating vision-specific information for both understanding and generation. Our experiments demonstrate that X-Fusion consistently outperforms alternative architectures on both image-to-text and text-to-image tasks. We find that incorporating understanding-focused data improves generation quality, reducing image data noise enhances overall performance, and feature alignment accelerates convergence for smaller models but has minimal impact on larger ones. Our findings provide valuable insights into building efficient unified multimodal models.
High-resolution myelin-water fraction and quantitative relaxation mapping using 3D ViSTa-MR fingerprinting
Dec 21, 2023Abstract:Purpose: This study aims to develop a high-resolution whole-brain multi-parametric quantitative MRI approach for simultaneous mapping of myelin-water fraction (MWF), T1, T2, and proton-density (PD), all within a clinically feasible scan time. Methods: We developed 3D ViSTa-MRF, which combined Visualization of Short Transverse relaxation time component (ViSTa) technique with MR Fingerprinting (MRF), to achieve high-fidelity whole-brain MWF and T1/T2/PD mapping on a clinical 3T scanner. To achieve fast acquisition and memory-efficient reconstruction, the ViSTa-MRF sequence leverages an optimized 3D tiny-golden-angle-shuffling spiral-projection acquisition and joint spatial-temporal subspace reconstruction with optimized preconditioning algorithm. With the proposed ViSTa-MRF approach, high-fidelity direct MWF mapping was achieved without a need for multi-compartment fitting that could introduce bias and/or noise from additional assumptions or priors. Results: The in-vivo results demonstrate the effectiveness of the proposed acquisition and reconstruction framework to provide fast multi-parametric mapping with high SNR and good quality. The in-vivo results of 1mm- and 0.66mm-iso datasets indicate that the MWF values measured by the proposed method are consistent with standard ViSTa results that are 30x slower with lower SNR. Furthermore, we applied the proposed method to enable 5-minute whole-brain 1mm-iso assessment of MWF and T1/T2/PD mappings for infant brain development and for post-mortem brain samples. Conclusions: In this work, we have developed a 3D ViSTa-MRF technique that enables the acquisition of whole-brain MWF, quantitative T1, T2, and PD maps at 1mm and 0.66mm isotropic resolution in 5 and 15 minutes, respectively. This advancement allows for quantitative investigations of myelination changes in the brain.
Optimized multi-axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole-brain high-isotropic-resolution quantitative imaging
Aug 12, 2021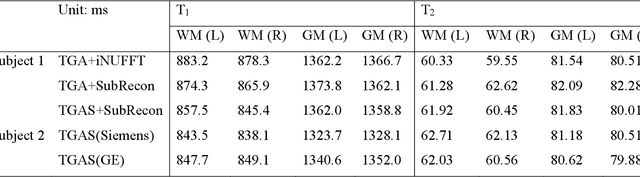
Abstract:Purpose: To improve image quality and accelerate the acquisition of 3D MRF. Methods: Building on the multi-axis spiral-projection MRF technique, a subspace reconstruction with locally low rank (LLR) constraint and a modified spiral-projection spatiotemporal encoding scheme termed tiny-golden-angle-shuffling (TGAS) were implemented for rapid whole-brain high-resolution quantitative mapping. The LLR regularization parameter and the number of subspace bases were tuned using retrospective in-vivo data and simulated examinations, respectively. B0 inhomogeneity correction using multi-frequency interpolation was incorporated into the subspace reconstruction to further improve the image quality by mitigating blurring caused by off-resonance effect. Results: The proposed MRF acquisition and reconstruction framework can produce provide high quality 1-mm isotropic whole-brain quantitative maps in a total acquisition time of 1 minute 55 seconds, with higher-quality results than ones obtained from the previous approach in 6 minutes. The comparison of quantitative results indicates that neither the subspace reconstruction nor the TGAS trajectory induce bias for T1 and T2 mapping. High quality whole-brain MRF data were also obtained at 0.66-mm isotropic resolution in 4 minutes using the proposed technique, where the increased resolution was shown to improve visualization of subtle brain structures. Conclusion: The proposed TGAS-SPI-MRF with optimized spiral-projection trajectory and subspace reconstruction can enable high-resolution quantitative mapping with faster acquisition speed.
Nonlinear Dipole Inversion (NDI) enables Quantitative Susceptibility Mapping (QSM) without parameter tuning
Sep 30, 2019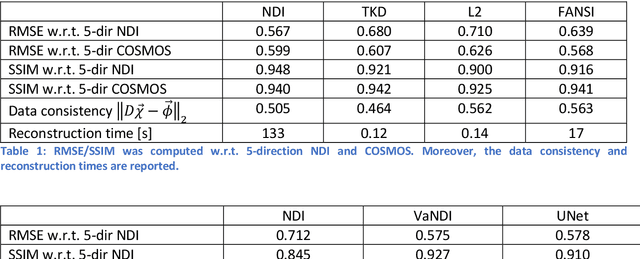
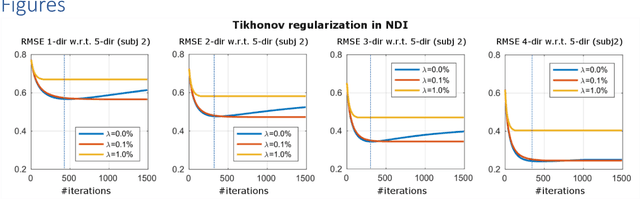
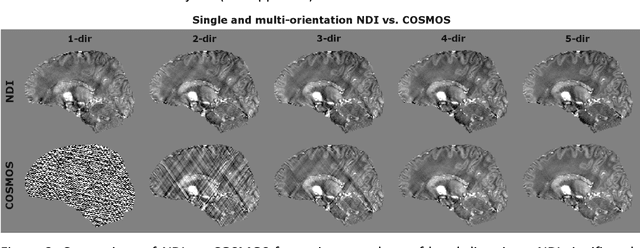
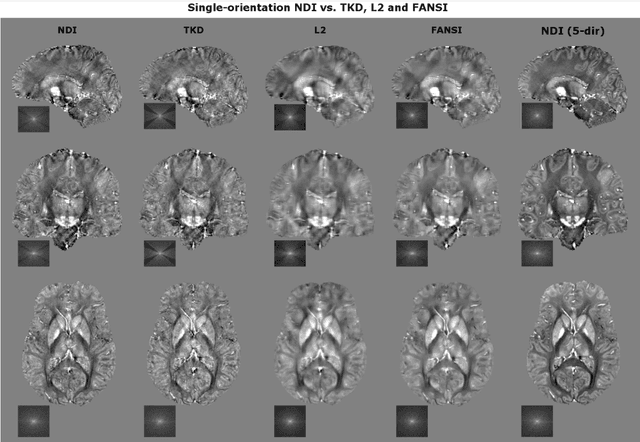
Abstract:We propose Nonlinear Dipole Inversion (NDI) for high-quality Quantitative Susceptibility Mapping (QSM) without regularization tuning, while matching the image quality of state-of-the-art reconstruction techniques. In addition to avoiding over-smoothing that these techniques often suffer from, we also obviate the need for parameter selection. NDI is flexible enough to allow for reconstruction from an arbitrary number of head orientations, and outperforms COSMOS even when using as few as 1-direction data. This is made possible by a nonlinear forward-model that uses the magnitude as an effective prior, for which we derived a simple gradient descent update rule. We synergistically combine this physics-model with a Variational Network (VN) to leverage the power of deep learning in the VaNDI algorithm. This technique adopts the simple gradient descent rule from NDI and learns the network parameters during training, hence requires no additional parameter tuning. Further, we evaluate NDI at 7T using highly accelerated Wave-CAIPI acquisitions at 0.5 mm isotropic resolution and demonstrate high-quality QSM from as few as 2-direction data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge