Hongjian He
Self-Supervised Elimination of Non-Independent Noise in Hyperspectral Imaging
Sep 16, 2024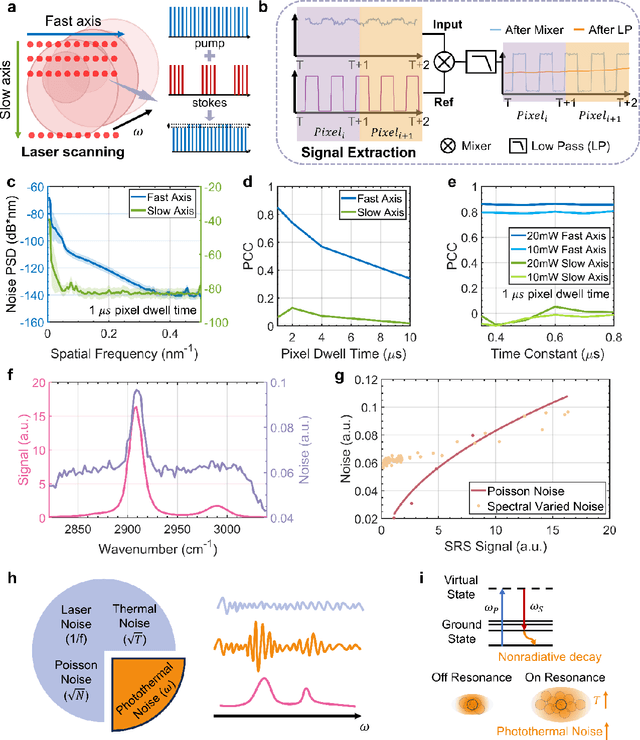
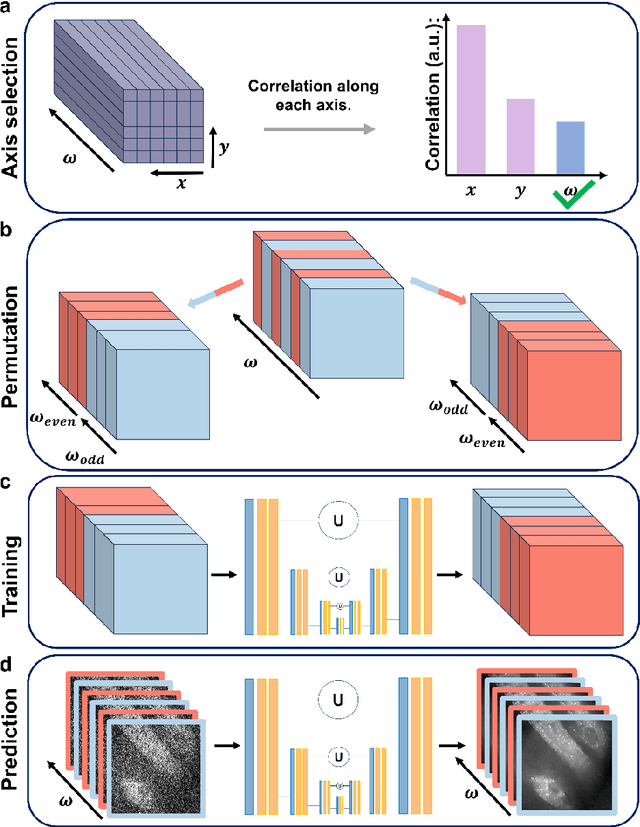
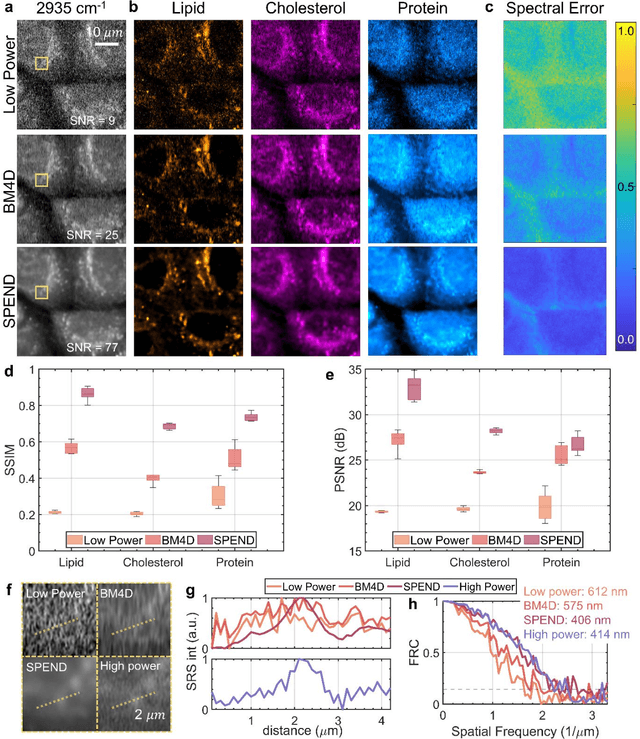
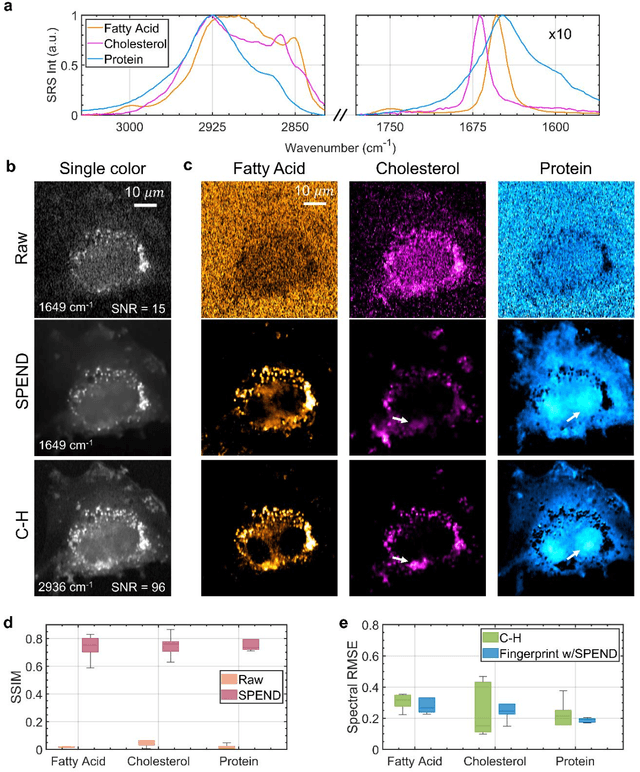
Abstract:Hyperspectral imaging has been widely used for spectral and spatial identification of target molecules, yet often contaminated by sophisticated noise. Current denoising methods generally rely on independent and identically distributed noise statistics, showing corrupted performance for non-independent noise removal. Here, we demonstrate Self-supervised PErmutation Noise2noise Denoising (SPEND), a deep learning denoising architecture tailor-made for removing non-independent noise from a single hyperspectral image stack. We utilize hyperspectral stimulated Raman scattering and mid-infrared photothermal microscopy as the testbeds, where the noise is spatially correlated and spectrally varied. Based on single hyperspectral images, SPEND permutates odd and even spectral frames to generate two stacks with identical noise properties, and uses the pairs for efficient self-supervised noise-to-noise training. SPEND achieved an 8-fold signal-to-noise improvement without having access to the ground truth data. SPEND enabled accurate mapping of low concentration biomolecules in both fingerprint and silent regions, demonstrating its robustness in sophisticated cellular environments.
High-resolution myelin-water fraction and quantitative relaxation mapping using 3D ViSTa-MR fingerprinting
Dec 21, 2023Abstract:Purpose: This study aims to develop a high-resolution whole-brain multi-parametric quantitative MRI approach for simultaneous mapping of myelin-water fraction (MWF), T1, T2, and proton-density (PD), all within a clinically feasible scan time. Methods: We developed 3D ViSTa-MRF, which combined Visualization of Short Transverse relaxation time component (ViSTa) technique with MR Fingerprinting (MRF), to achieve high-fidelity whole-brain MWF and T1/T2/PD mapping on a clinical 3T scanner. To achieve fast acquisition and memory-efficient reconstruction, the ViSTa-MRF sequence leverages an optimized 3D tiny-golden-angle-shuffling spiral-projection acquisition and joint spatial-temporal subspace reconstruction with optimized preconditioning algorithm. With the proposed ViSTa-MRF approach, high-fidelity direct MWF mapping was achieved without a need for multi-compartment fitting that could introduce bias and/or noise from additional assumptions or priors. Results: The in-vivo results demonstrate the effectiveness of the proposed acquisition and reconstruction framework to provide fast multi-parametric mapping with high SNR and good quality. The in-vivo results of 1mm- and 0.66mm-iso datasets indicate that the MWF values measured by the proposed method are consistent with standard ViSTa results that are 30x slower with lower SNR. Furthermore, we applied the proposed method to enable 5-minute whole-brain 1mm-iso assessment of MWF and T1/T2/PD mappings for infant brain development and for post-mortem brain samples. Conclusions: In this work, we have developed a 3D ViSTa-MRF technique that enables the acquisition of whole-brain MWF, quantitative T1, T2, and PD maps at 1mm and 0.66mm isotropic resolution in 5 and 15 minutes, respectively. This advancement allows for quantitative investigations of myelination changes in the brain.
Unified Style Transfer
Oct 20, 2021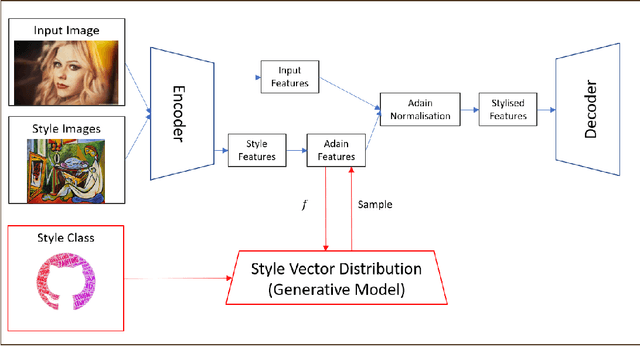
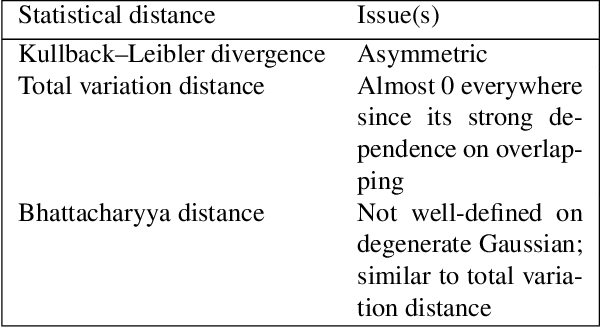
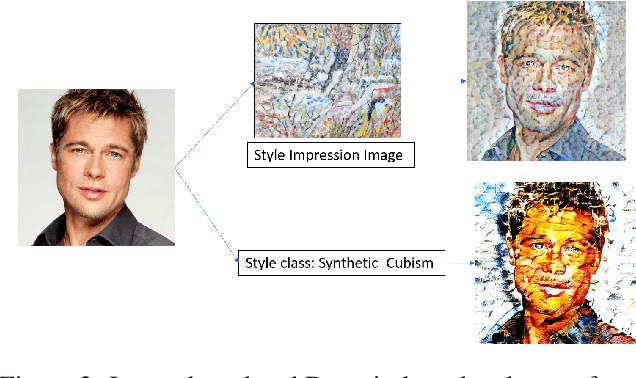
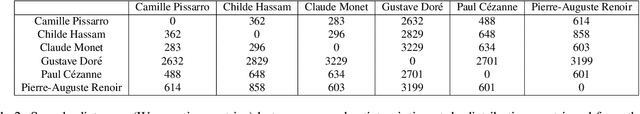
Abstract:Currently, it is hard to compare and evaluate different style transfer algorithms due to chaotic definitions of style and the absence of agreed objective validation methods in the study of style transfer. In this paper, a novel approach, the Unified Style Transfer (UST) model, is proposed. With the introduction of a generative model for internal style representation, UST can transfer images in two approaches, i.e., Domain-based and Image-based, simultaneously. At the same time, a new philosophy based on the human sense of art and style distributions for evaluating the transfer model is presented and demonstrated, called Statistical Style Analysis. It provides a new path to validate style transfer models' feasibility by validating the general consistency between internal style representation and art facts. Besides, the translation-invariance of AdaIN features is also discussed.
Optimized multi-axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole-brain high-isotropic-resolution quantitative imaging
Aug 12, 2021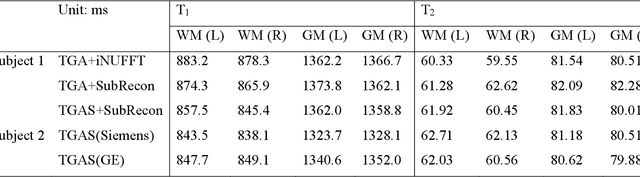
Abstract:Purpose: To improve image quality and accelerate the acquisition of 3D MRF. Methods: Building on the multi-axis spiral-projection MRF technique, a subspace reconstruction with locally low rank (LLR) constraint and a modified spiral-projection spatiotemporal encoding scheme termed tiny-golden-angle-shuffling (TGAS) were implemented for rapid whole-brain high-resolution quantitative mapping. The LLR regularization parameter and the number of subspace bases were tuned using retrospective in-vivo data and simulated examinations, respectively. B0 inhomogeneity correction using multi-frequency interpolation was incorporated into the subspace reconstruction to further improve the image quality by mitigating blurring caused by off-resonance effect. Results: The proposed MRF acquisition and reconstruction framework can produce provide high quality 1-mm isotropic whole-brain quantitative maps in a total acquisition time of 1 minute 55 seconds, with higher-quality results than ones obtained from the previous approach in 6 minutes. The comparison of quantitative results indicates that neither the subspace reconstruction nor the TGAS trajectory induce bias for T1 and T2 mapping. High quality whole-brain MRF data were also obtained at 0.66-mm isotropic resolution in 4 minutes using the proposed technique, where the increased resolution was shown to improve visualization of subtle brain structures. Conclusion: The proposed TGAS-SPI-MRF with optimized spiral-projection trajectory and subspace reconstruction can enable high-resolution quantitative mapping with faster acquisition speed.
Model-based Synthetic Data-driven Learning (MOST-DL): Application in Single-shot T2 Mapping with Severe Head Motion Using Overlapping-echo Acquisition
Jul 30, 2021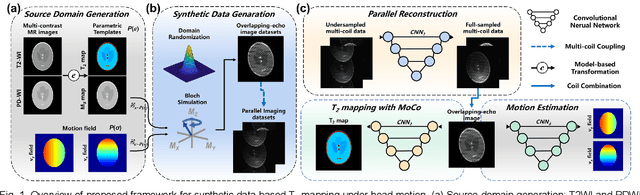
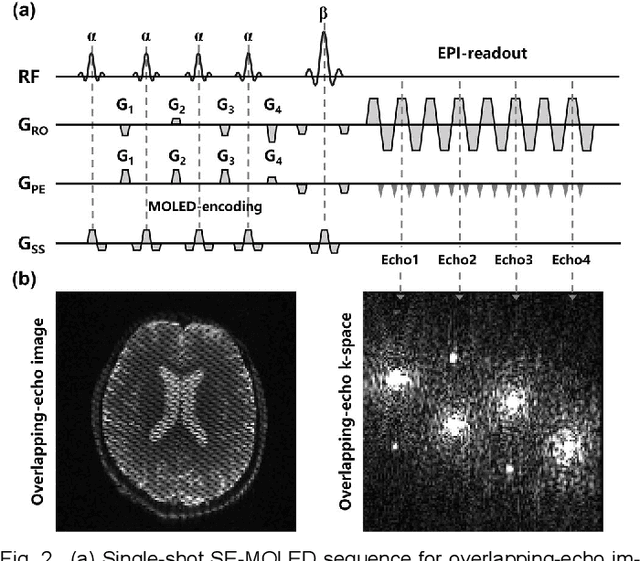
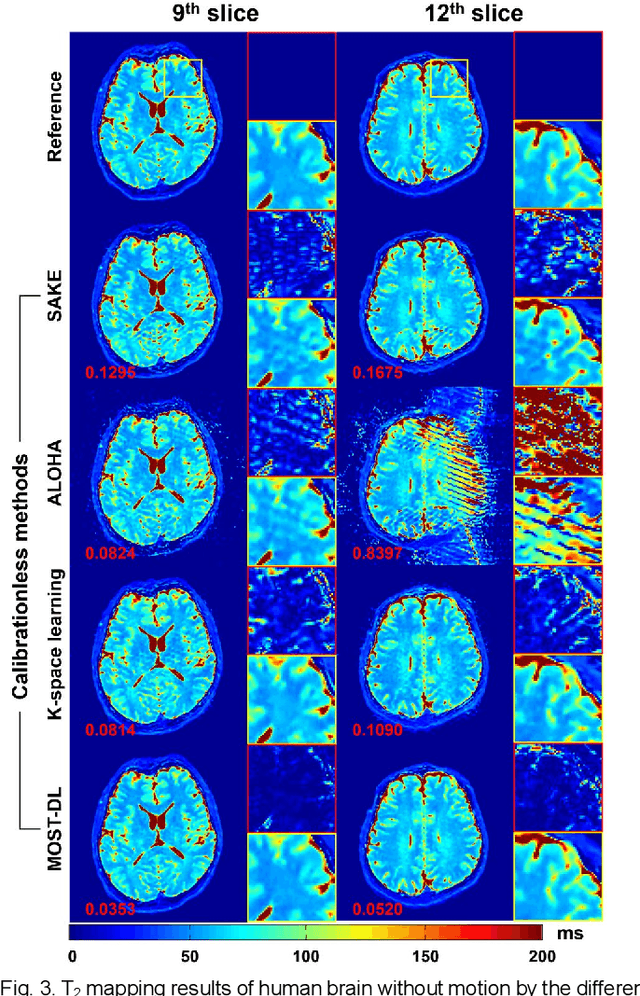
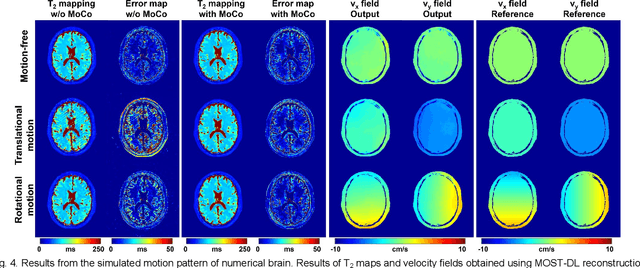
Abstract:Data-driven learning algorithm has been successfully applied to facilitate reconstruction of medical imaging. However, real-world data needed for supervised learning are typically unavailable or insufficient, especially in the field of magnetic resonance imaging (MRI). Synthetic training samples have provided a potential solution for such problem, while the challenge brought by various non-ideal situations were usually encountered especially under complex experimental conditions. In this study, a general framework, Model-based Synthetic Data-driven Learning (MOST-DL), was proposed to generate paring data for network training to achieve robust T2 mapping using overlapping-echo acquisition under severe head motion accompanied with inhomogeneous RF field. We decomposed this challenging task into parallel reconstruction and motion correction according to a forward model. The neural network was first trained in pure synthetic dataset and then evaluated with in vivo human brain. Experiments showed that MOST-DL method significantly reduces ghosting and motion artifacts in T2 maps in the presence of random and continuous subject movement. We believe that the proposed approach may open a door for solving similar problems with other MRI acquisition methods and can be extended to other areas of medical imaging.
Reconstruction of Quantitative Susceptibility Maps from Phase of Susceptibility Weighted Imaging with Cross-Connected Ψ-Net
Oct 14, 2020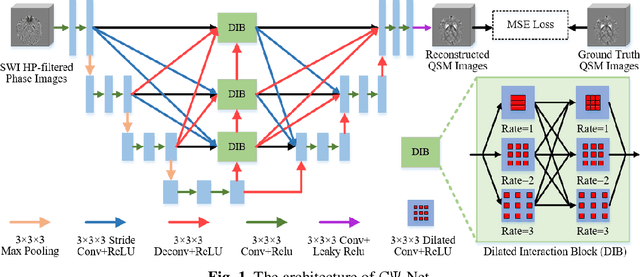
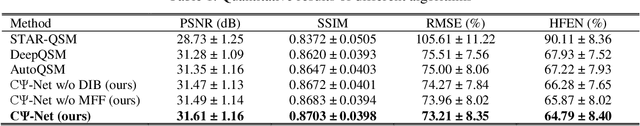
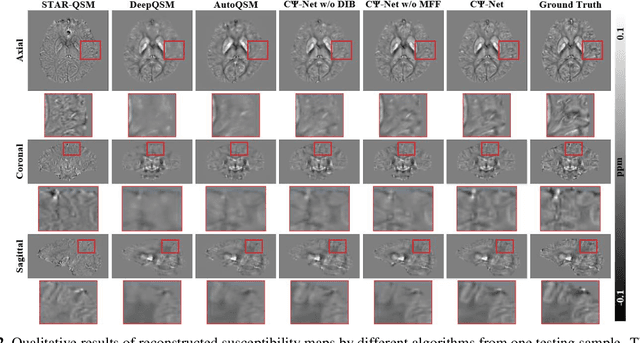
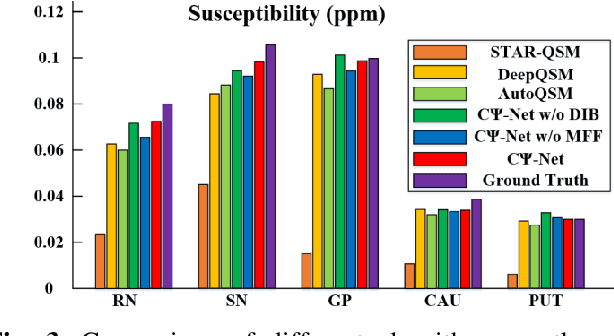
Abstract:Quantitative Susceptibility Mapping (QSM) is a new phase-based technique for quantifying magnetic susceptibility. The existing QSM reconstruction methods generally require complicated pre-processing on high-quality phase data. In this work, we propose to explore a new value of the high-pass filtered phase data generated in susceptibility weighted imaging (SWI), and develop an end-to-end Cross-connected {\Psi}-Net (C{\Psi}-Net) to reconstruct QSM directly from these phase data in SWI without additional pre-processing. C{\Psi}-Net adds an intermediate branch in the classical U-Net to form a {\Psi}-like structure. The specially designed dilated interaction block is embedded in each level of this branch to enlarge the receptive fields for capturing more susceptibility information from a wider spatial range of phase images. Moreover, the crossed connections are utilized between branches to implement a multi-resolution feature fusion scheme, which helps C{\Psi}-Net capture rich contextual information for accurate reconstruction. The experimental results on a human dataset show that C{\Psi}-Net achieves superior performance in our task over other QSM reconstruction algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge