Congbo Cai
Bridging Synthetic-to-Real Gaps: Frequency-Aware Perturbation and Selection for Single-shot Multi-Parametric Mapping Reconstruction
Mar 05, 2025Abstract:Data-centric artificial intelligence (AI) has remarkably advanced medical imaging, with emerging methods using synthetic data to address data scarcity while introducing synthetic-to-real gaps. Unsupervised domain adaptation (UDA) shows promise in ground truth-scarce tasks, but its application in reconstruction remains underexplored. Although multiple overlapping-echo detachment (MOLED) achieves ultra-fast multi-parametric reconstruction, extending its application to various clinical scenarios, the quality suffers from deficiency in mitigating the domain gap, difficulty in maintaining structural integrity, and inadequacy in ensuring mapping accuracy. To resolve these issues, we proposed frequency-aware perturbation and selection (FPS), comprising Wasserstein distance-modulated frequency-aware perturbation (WDFP) and hierarchical frequency-aware selection network (HFSNet), which integrates frequency-aware adaptive selection (FAS), compact FAS (cFAS) and feature-aware architecture integration (FAI). Specifically, perturbation activates domain-invariant feature learning within uncertainty, while selection refines optimal solutions within perturbation, establishing a robust and closed-loop learning pathway. Extensive experiments on synthetic data, along with diverse real clinical cases from 5 healthy volunteers, 94 ischemic stroke patients, and 46 meningioma patients, demonstrate the superiority and clinical applicability of FPS. Furthermore, FPS is applied to diffusion tensor imaging (DTI), underscoring its versatility and potential for broader medical applications. The code is available at https://github.com/flyannie/FPS.
MvKeTR: Chest CT Report Generation with Multi-View Perception and Knowledge Enhancement
Nov 27, 2024Abstract:CT report generation (CTRG) aims to automatically generate diagnostic reports for 3D volumes, relieving clinicians' workload and improving patient care. Despite clinical value, existing works fail to effectively incorporate diagnostic information from multiple anatomical views and lack related clinical expertise essential for accurate and reliable diagnosis. To resolve these limitations, we propose a novel Multi-view perception Knowledge-enhanced Tansformer (MvKeTR) to mimic the diagnostic workflow of clinicians. Just as radiologists first examine CT scans from multiple planes, a Multi-View Perception Aggregator (MVPA) with view-aware attention effectively synthesizes diagnostic information from multiple anatomical views. Then, inspired by how radiologists further refer to relevant clinical records to guide diagnostic decision-making, a Cross-Modal Knowledge Enhancer (CMKE) retrieves the most similar reports based on the query volume to incorporate domain knowledge into the diagnosis procedure. Furthermore, instead of traditional MLPs, we employ Kolmogorov-Arnold Networks (KANs) with learnable nonlinear activation functions as the fundamental building blocks of both modules to better capture intricate diagnostic patterns in CT interpretation. Extensive experiments on the public CTRG-Chest-548K dataset demonstrate that our method outpaces prior state-of-the-art models across all metrics.
Simultaneous Deep Learning of Myocardium Segmentation and T2 Quantification for Acute Myocardial Infarction MRI
May 17, 2024



Abstract:In cardiac Magnetic Resonance Imaging (MRI) analysis, simultaneous myocardial segmentation and T2 quantification are crucial for assessing myocardial pathologies. Existing methods often address these tasks separately, limiting their synergistic potential. To address this, we propose SQNet, a dual-task network integrating Transformer and Convolutional Neural Network (CNN) components. SQNet features a T2-refine fusion decoder for quantitative analysis, leveraging global features from the Transformer, and a segmentation decoder with multiple local region supervision for enhanced accuracy. A tight coupling module aligns and fuses CNN and Transformer branch features, enabling SQNet to focus on myocardium regions. Evaluation on healthy controls (HC) and acute myocardial infarction patients (AMI) demonstrates superior segmentation dice scores (89.3/89.2) compared to state-of-the-art methods (87.7/87.9). T2 quantification yields strong linear correlations (Pearson coefficients: 0.84/0.93) with label values for HC/AMI, indicating accurate mapping. Radiologist evaluations confirm SQNet's superior image quality scores (4.60/4.58 for segmentation, 4.32/4.42 for T2 quantification) over state-of-the-art methods (4.50/4.44 for segmentation, 3.59/4.37 for T2 quantification). SQNet thus offers accurate simultaneous segmentation and quantification, enhancing cardiac disease diagnosis, such as AMI.
Deep Separable Spatiotemporal Learning for Fast Dynamic Cardiac MRI
Feb 24, 2024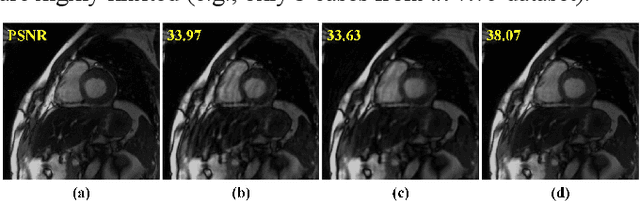
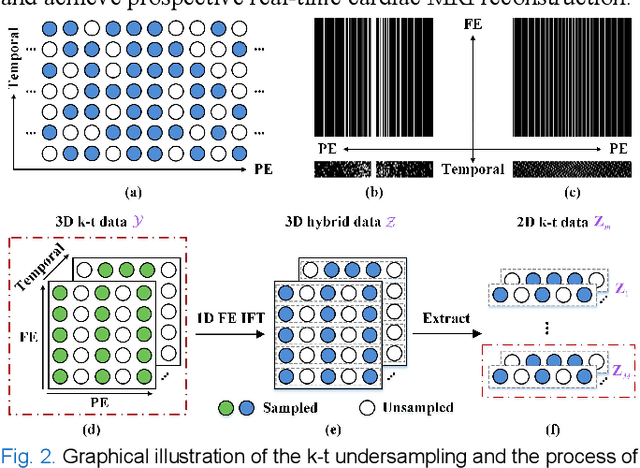
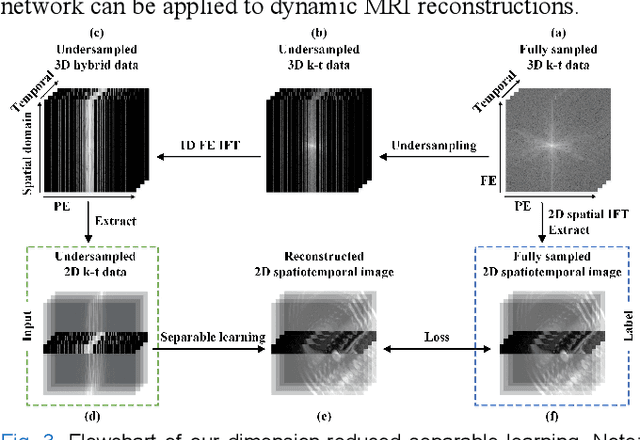
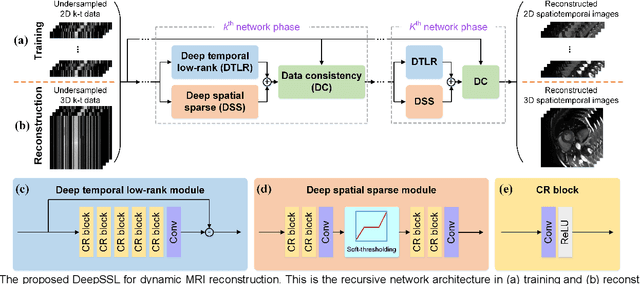
Abstract:Dynamic magnetic resonance imaging (MRI) plays an indispensable role in cardiac diagnosis. To enable fast imaging, the k-space data can be undersampled but the image reconstruction poses a great challenge of high-dimensional processing. This challenge leads to necessitate extensive training data in many deep learning reconstruction methods. This work proposes a novel and efficient approach, leveraging a dimension-reduced separable learning scheme that excels even with highly limited training data. We further integrate it with spatiotemporal priors to develop a Deep Separable Spatiotemporal Learning network (DeepSSL), which unrolls an iteration process of a reconstruction model with both temporal low-rankness and spatial sparsity. Intermediate outputs are visualized to provide insights into the network's behavior and enhance its interpretability. Extensive results on cardiac cine datasets show that the proposed DeepSSL is superior to the state-of-the-art methods visually and quantitatively, while reducing the demand for training cases by up to 75%. And its preliminary adaptability to cardiac patients has been verified through experienced radiologists' and cardiologists' blind reader study. Additionally, DeepSSL also benefits for achieving the downstream task of cardiac segmentation with higher accuracy and shows robustness in prospective real-time cardiac MRI.
High-efficient deep learning-based DTI reconstruction with flexible diffusion gradient encoding scheme
Aug 02, 2023Abstract:Purpose: To develop and evaluate a novel dynamic-convolution-based method called FlexDTI for high-efficient diffusion tensor reconstruction with flexible diffusion encoding gradient schemes. Methods: FlexDTI was developed to achieve high-quality DTI parametric mapping with flexible number and directions of diffusion encoding gradients. The proposed method used dynamic convolution kernels to embed diffusion gradient direction information into feature maps of the corresponding diffusion signal. Besides, our method realized the generalization of a flexible number of diffusion gradient directions by setting the maximum number of input channels of the network. The network was trained and tested using data sets from the Human Connectome Project and a local hospital. Results from FlexDTI and other advanced tensor parameter estimation methods were compared. Results: Compared to other methods, FlexDTI successfully achieves high-quality diffusion tensor-derived variables even if the number and directions of diffusion encoding gradients are variable. It increases peak signal-to-noise ratio (PSNR) by about 10 dB on Fractional Anisotropy (FA) and Mean Diffusivity (MD), compared with the state-of-the-art deep learning method with flexible diffusion encoding gradient schemes. Conclusion: FlexDTI can well learn diffusion gradient direction information to achieve generalized DTI reconstruction with flexible diffusion gradient schemes. Both flexibility and reconstruction quality can be taken into account in this network.
One for Multiple: Physics-informed Synthetic Data Boosts Generalizable Deep Learning for Fast MRI Reconstruction
Jul 25, 2023
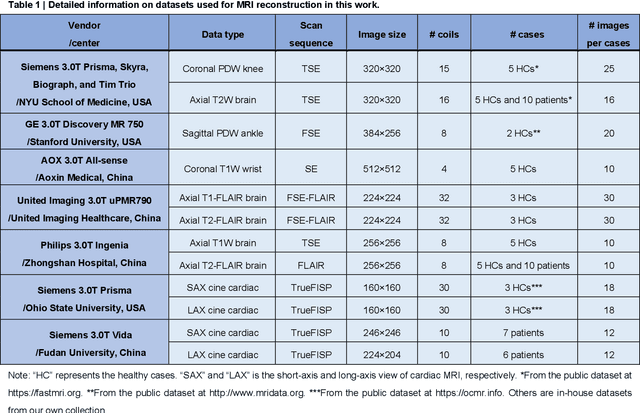
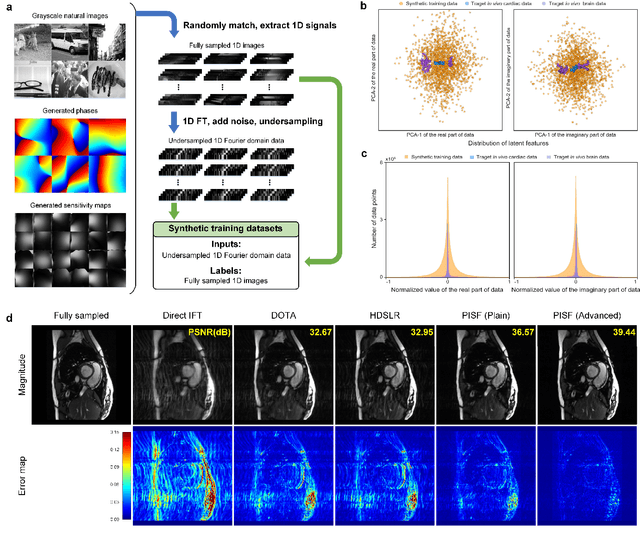
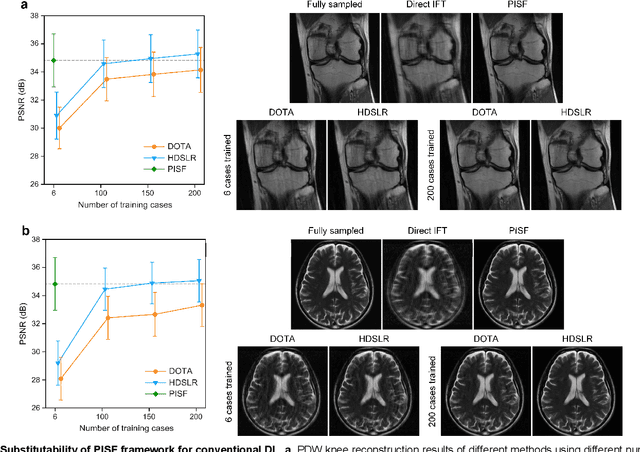
Abstract:Magnetic resonance imaging (MRI) is a principal radiological modality that provides radiation-free, abundant, and diverse information about the whole human body for medical diagnosis, but suffers from prolonged scan time. The scan time can be significantly reduced through k-space undersampling but the introduced artifacts need to be removed in image reconstruction. Although deep learning (DL) has emerged as a powerful tool for image reconstruction in fast MRI, its potential in multiple imaging scenarios remains largely untapped. This is because not only collecting large-scale and diverse realistic training data is generally costly and privacy-restricted, but also existing DL methods are hard to handle the practically inevitable mismatch between training and target data. Here, we present a Physics-Informed Synthetic data learning framework for Fast MRI, called PISF, which is the first to enable generalizable DL for multi-scenario MRI reconstruction using solely one trained model. For a 2D image, the reconstruction is separated into many 1D basic problems and starts with the 1D data synthesis, to facilitate generalization. We demonstrate that training DL models on synthetic data, integrated with enhanced learning techniques, can achieve comparable or even better in vivo MRI reconstruction compared to models trained on a matched realistic dataset, reducing the demand for real-world MRI data by up to 96%. Moreover, our PISF shows impressive generalizability in multi-vendor multi-center imaging. Its excellent adaptability to patients has been verified through 10 experienced doctors' evaluations. PISF provides a feasible and cost-effective way to markedly boost the widespread usage of DL in various fast MRI applications, while freeing from the intractable ethical and practical considerations of in vivo human data acquisitions.
High-efficient Bloch simulation of magnetic resonance imaging sequences based on deep learning
Oct 19, 2022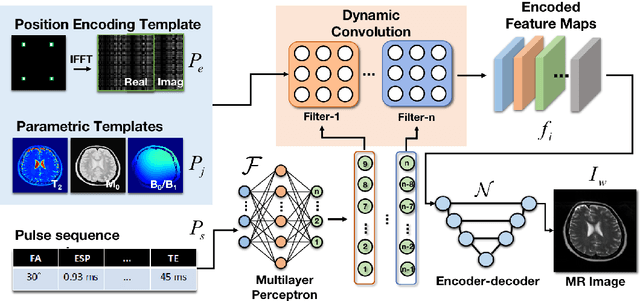

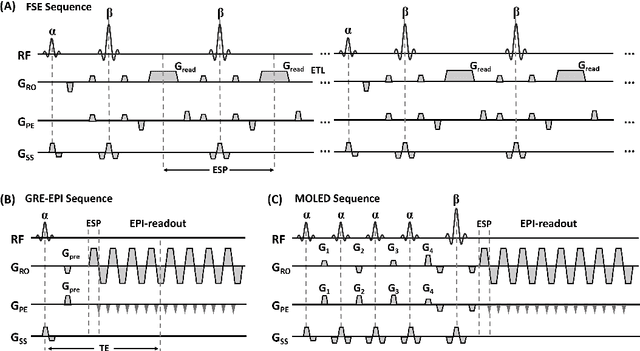

Abstract:Bloch simulation constitutes an essential part of magnetic resonance imaging (MRI) development. However, even with the graphics processing units (GPU) acceleration, the heavy computational load remains a major challenge, especially in large-scale, high-accuracy simulation scenarios. Here we present a framework based on convolutional neural networks to build a high-efficient 2D Bloch simulator, termed Simu-Net. Compared to the mainstream GPU-based MRI simulation software, Simu-Net successfully accelerates simulations by over hundreds of times in three MRI pulse sequences. The accuracy and robustness of the proposed framework were also verified qualitatively and quantitatively. The trained Simu-Net was applied to generate sufficient customized training samples for deep learning-based T2 mapping and comparable results to conventional methods were obtained in the human brain. As a proof-of-concept work, Simu-Net shows the potential to apply deep learning for rapidly approximating the Bloch equation as a forward physical process.
Physics-driven Synthetic Data Learning for Biomedical Magnetic Resonance
Mar 22, 2022



Abstract:Deep learning has innovated the field of computational imaging. One of its bottlenecks is unavailable or insufficient training data. This article reviews an emerging paradigm, imaging physics-based data synthesis (IPADS), that can provide huge training data in biomedical magnetic resonance without or with few real data. Following the physical law of magnetic resonance, IPADS generates signals from differential equations or analytical solution models, making the learning more scalable, explainable, and better protecting privacy. Key components of IPADS learning, including signal generation models, basic deep learning network structures, enhanced data generation, and learning methods are discussed. Great potentials of IPADS have been demonstrated by representative applications in fast imaging, ultrafast signal reconstruction and accurate parameter quantification. Finally, open questions and future work have been discussed.
Model-based Synthetic Data-driven Learning (MOST-DL): Application in Single-shot T2 Mapping with Severe Head Motion Using Overlapping-echo Acquisition
Jul 30, 2021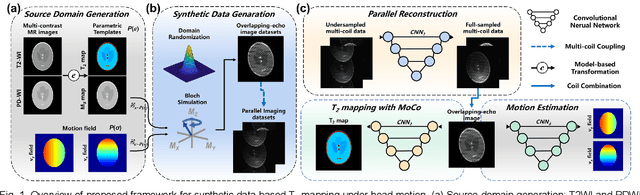
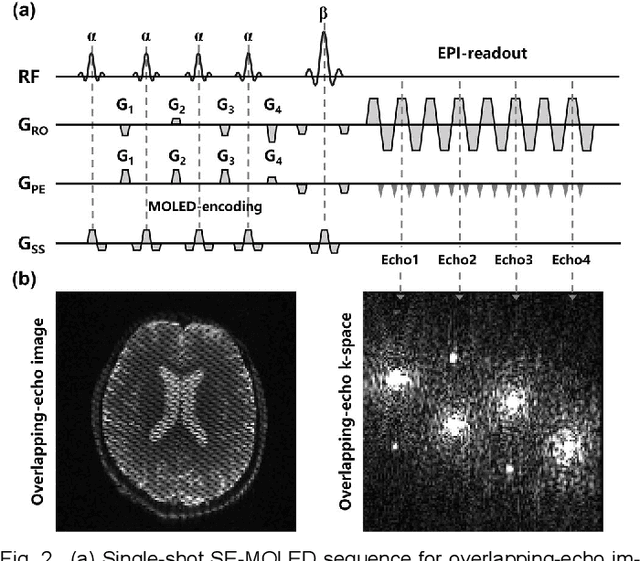
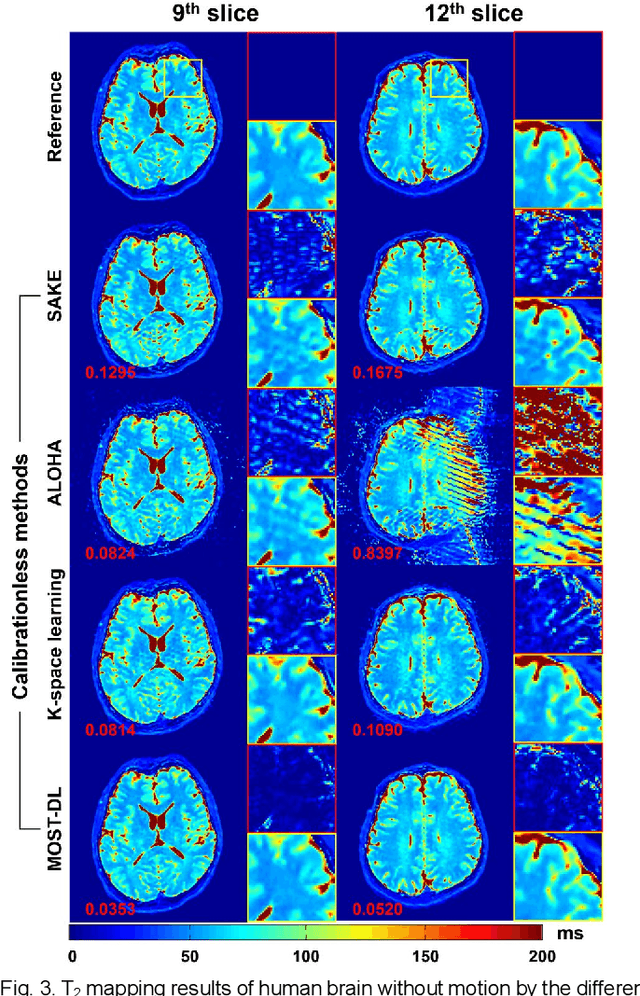
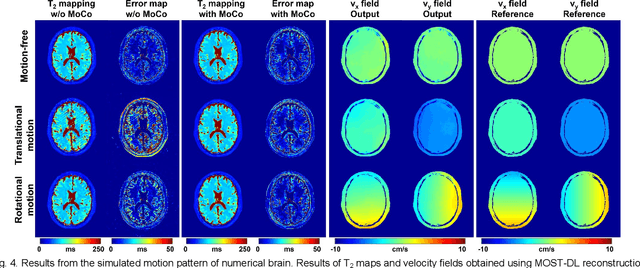
Abstract:Data-driven learning algorithm has been successfully applied to facilitate reconstruction of medical imaging. However, real-world data needed for supervised learning are typically unavailable or insufficient, especially in the field of magnetic resonance imaging (MRI). Synthetic training samples have provided a potential solution for such problem, while the challenge brought by various non-ideal situations were usually encountered especially under complex experimental conditions. In this study, a general framework, Model-based Synthetic Data-driven Learning (MOST-DL), was proposed to generate paring data for network training to achieve robust T2 mapping using overlapping-echo acquisition under severe head motion accompanied with inhomogeneous RF field. We decomposed this challenging task into parallel reconstruction and motion correction according to a forward model. The neural network was first trained in pure synthetic dataset and then evaluated with in vivo human brain. Experiments showed that MOST-DL method significantly reduces ghosting and motion artifacts in T2 maps in the presence of random and continuous subject movement. We believe that the proposed approach may open a door for solving similar problems with other MRI acquisition methods and can be extended to other areas of medical imaging.
A Segmentation-aware Deep Fusion Network for Compressed Sensing MRI
Apr 04, 2018



Abstract:Compressed sensing MRI is a classic inverse problem in the field of computational imaging, accelerating the MR imaging by measuring less k-space data. The deep neural network models provide the stronger representation ability and faster reconstruction compared with "shallow" optimization-based methods. However, in the existing deep-based CS-MRI models, the high-level semantic supervision information from massive segmentation-labels in MRI dataset is overlooked. In this paper, we proposed a segmentation-aware deep fusion network called SADFN for compressed sensing MRI. The multilayer feature aggregation (MLFA) method is introduced here to fuse all the features from different layers in the segmentation network. Then, the aggregated feature maps containing semantic information are provided to each layer in the reconstruction network with a feature fusion strategy. This guarantees the reconstruction network is aware of the different regions in the image it reconstructs, simplifying the function mapping. We prove the utility of the cross-layer and cross-task information fusion strategy by comparative study. Extensive experiments on brain segmentation benchmark MRBrainS validated that the proposed SADFN model achieves state-of-the-art accuracy in compressed sensing MRI. This paper provides a novel approach to guide the low-level visual task using the information from mid- or high-level task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge