Liyan Sun
Exploring Siamese Networks in Self-Supervised Fast MRI Reconstruction
Jan 18, 2025



Abstract:Reconstructing MR images using deep neural networks from undersampled k-space data without using fully sampled training references offers significant value in practice, which is a self-supervised regression problem calling for effective prior knowledge and supervision. The Siamese architectures are motivated by the definition "invariance" and shows promising results in unsupervised visual representative learning. Building homologous transformed images and avoiding trivial solutions are two major challenges in Siamese-based self-supervised model. In this work, we explore Siamese architecture for MRI reconstruction in a self-supervised training fashion called SiamRecon. We show the proposed approach mimics an expectation maximization algorithm. The alternative optimization provide effective supervision signal and avoid collapse. The proposed SiamRecon achieves the state-of-the-art reconstruction accuracy in the field of self-supervised learning on both single-coil brain MRI and multi-coil knee MRI.
A Closer Look at Personalization in Federated Image Classification
Apr 22, 2022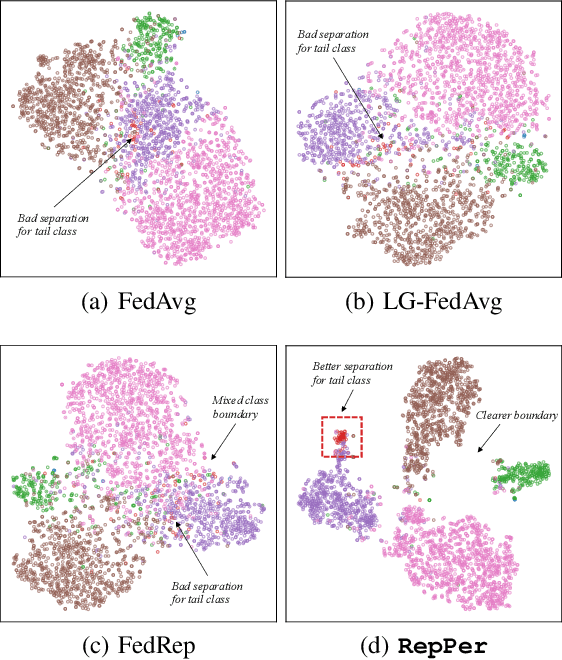
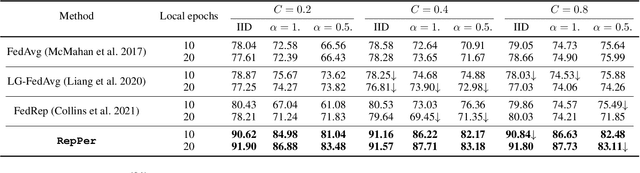

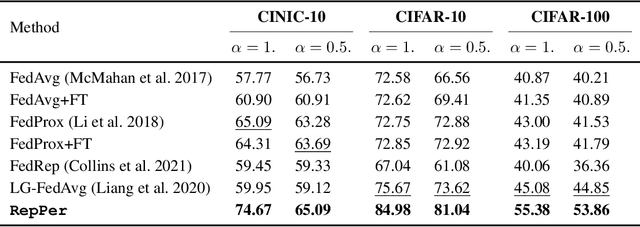
Abstract:Federated Learning (FL) is developed to learn a single global model across the decentralized data, while is susceptible when realizing client-specific personalization in the presence of statistical heterogeneity. However, studies focus on learning a robust global model or personalized classifiers, which yield divergence due to inconsistent objectives. This paper shows that it is possible to achieve flexible personalization after the convergence of the global model by introducing representation learning. In this paper, we first analyze and determine that non-IID data harms representation learning of the global model. Existing FL methods adhere to the scheme of jointly learning representations and classifiers, where the global model is an average of classification-based local models that are consistently subject to heterogeneity from non-IID data. As a solution, we separate representation learning from classification learning in FL and propose RepPer, an independent two-stage personalized FL framework.We first learn the client-side feature representation models that are robust to non-IID data and aggregate them into a global common representation model. After that, we achieve personalization by learning a classifier head for each client, based on the common representation obtained at the former stage. Notably, the proposed two-stage learning scheme of RepPer can be potentially used for lightweight edge computing that involves devices with constrained computation power.Experiments on various datasets (CIFAR-10/100, CINIC-10) and heterogeneous data setup show that RepPer outperforms alternatives in flexibility and personalization on non-IID data.
AFSC: Adaptive Fourier Space Compression for Anomaly Detection
Apr 17, 2022



Abstract:Anomaly Detection (AD) on medical images enables a model to recognize any type of anomaly pattern without lesion-specific supervised learning. Data augmentation based methods construct pseudo-healthy images by "pasting" fake lesions on real healthy ones, and a network is trained to predict healthy images in a supervised manner. The lesion can be found by difference between the unhealthy input and pseudo-healthy output. However, using only manually designed fake lesions fail to approximate to irregular real lesions, hence limiting the model generalization. We assume by exploring the intrinsic data property within images, we can distinguish previously unseen lesions from healthy regions in an unhealthy image. In this study, we propose an Adaptive Fourier Space Compression (AFSC) module to distill healthy feature for AD. The compression of both magnitude and phase in frequency domain addresses the hyper intensity and diverse position of lesions. Experimental results on the BraTS and MS-SEG datasets demonstrate an AFSC baseline is able to produce promising detection results, and an AFSC module can be effectively embedded into existing AD methods.
Hierarchical Deep Network with Uncertainty-aware Semi-supervised Learning for Vessel Segmentation
May 31, 2021



Abstract:The analysis of organ vessels is essential for computer-aided diagnosis and surgical planning. But it is not a easy task since the fine-detailed connected regions of organ vessel bring a lot of ambiguity in vessel segmentation and sub-type recognition, especially for the low-contrast capillary regions. Furthermore, recent two-staged approaches would accumulate and even amplify these inaccuracies from the first-stage whole vessel segmentation into the second-stage sub-type vessel pixel-wise classification. Moreover, the scarcity of manual annotation in organ vessels poses another challenge. In this paper, to address the above issues, we propose a hierarchical deep network where an attention mechanism localizes the low-contrast capillary regions guided by the whole vessels, and enhance the spatial activation in those areas for the sub-type vessels. In addition, we propose an uncertainty-aware semi-supervised training framework to alleviate the annotation-hungry limitation of deep models. The proposed method achieves the state-of-the-art performance in the benchmarks of both retinal artery/vein segmentation in fundus images and liver portal/hepatic vessel segmentation in CT images.
Few-shot Medical Image Segmentation using a Global Correlation Network with Discriminative Embedding
Dec 10, 2020



Abstract:Despite deep convolutional neural networks achieved impressive progress in medical image computing and analysis, its paradigm of supervised learning demands a large number of annotations for training to avoid overfitting and achieving promising results. In clinical practices, massive semantic annotations are difficult to acquire in some conditions where specialized biomedical expert knowledge is required, and it is also a common condition where only few annotated classes are available. In this work, we proposed a novel method for few-shot medical image segmentation, which enables a segmentation model to fast generalize to an unseen class with few training images. We construct our few-shot image segmentor using a deep convolutional network trained episodically. Motivated by the spatial consistency and regularity in medical images, we developed an efficient global correlation module to capture the correlation between a support and query image and incorporate it into the deep network called global correlation network. Moreover, we enhance discriminability of deep embedding to encourage clustering of the feature domains of the same class while keep the feature domains of different organs far apart. Ablation Study proved the effectiveness of the proposed global correlation module and discriminative embedding loss. Extensive experiments on anatomical abdomen images on both CT and MRI modalities are performed to demonstrate the state-of-the-art performance of our proposed model.
A Teacher-Student Framework for Semi-supervised Medical Image Segmentation From Mixed Supervision
Oct 23, 2020



Abstract:Standard segmentation of medical images based on full-supervised convolutional networks demands accurate dense annotations. Such learning framework is built on laborious manual annotation with restrict demands for expertise, leading to insufficient high-quality labels. To overcome such limitation and exploit massive weakly labeled data, we relaxed the rigid labeling requirement and developed a semi-supervised learning framework based on a teacher-student fashion for organ and lesion segmentation with partial dense-labeled supervision and supplementary loose bounding-box supervision which are easier to acquire. Observing the geometrical relation of an organ and its inner lesions in most cases, we propose a hierarchical organ-to-lesion (O2L) attention module in a teacher segmentor to produce pseudo-labels. Then a student segmentor is trained with combinations of manual-labeled and pseudo-labeled annotations. We further proposed a localization branch realized via an aggregation of high-level features in a deep decoder to predict locations of organ and lesion, which enriches student segmentor with precise localization information. We validated each design in our model on LiTS challenge datasets by ablation study and showed its state-of-the-art performance compared with recent methods. We show our model is robust to the quality of bounding box and achieves comparable performance compared with full-supervised learning methods.
An Adversarial Learning Approach to Medical Image Synthesis for Lesion Removal
Oct 25, 2018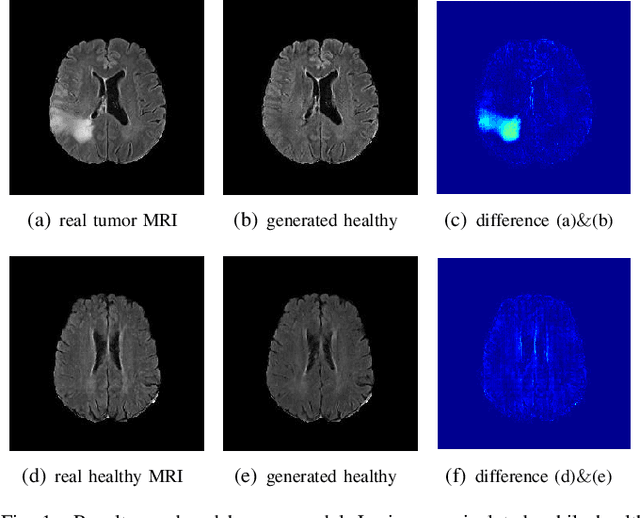
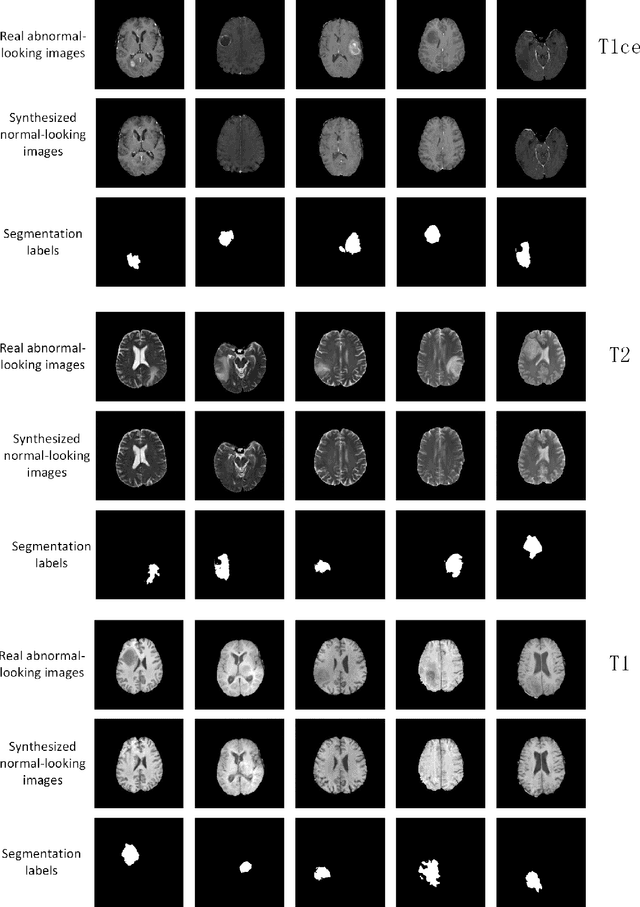
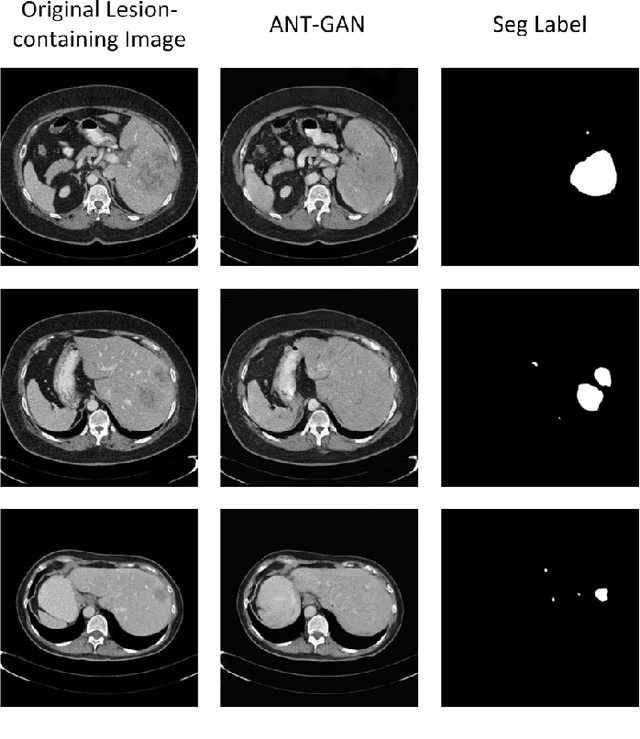
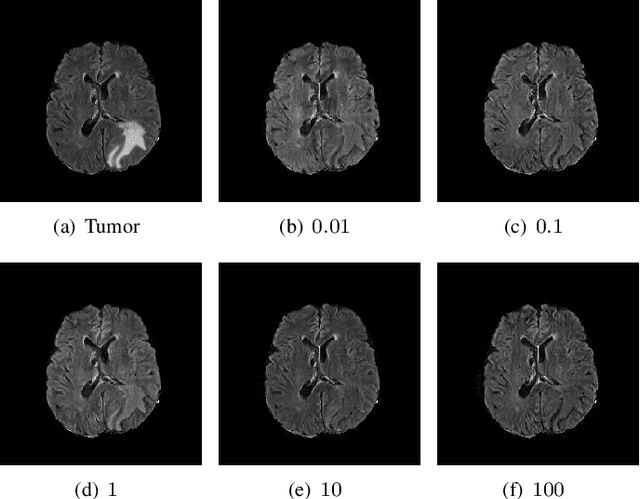
Abstract:The analysis of lesion within medical image data is desirable for efficient disease diagnosis, treatment and prognosis. The common lesion analysis tasks like segmentation and classification are mainly based on supervised learning with well-paired image-level or voxel-level labels. However, labeling the lesion in medical images is laborious requiring highly specialized knowledge. Inspired by the fact that radiologists make diagnoses based on expert knowledge on "healthiness" and "unhealthiness" developed from extensive experience, we propose an medical image synthesis model named abnormal-to-normal translation generative adversarial network (ANT-GAN) to predict a normal-looking medical image based on its abnormal-looking counterpart without the need of paired data for training. Unlike typical GANs, whose aim is to generate realistic samples with variations, our more restrictive model aims at producing the underlying normal-looking image corresponding to an image containing lesions, and thus requires a specialized design. With an ability to segment normal from abnormal tissue, our model is able to generate a highly realistic lesion-free medical image based on its true lesion-containing counterpart. Being able to provide a "normal" version of a medical image (possibly the same image if there is no illness) is not only an intriguing topic, but also can serve as a pre-processing and provide useful side information for medical imaging tasks like lesion segmentation or classification validated by our experiments.
Joint CS-MRI Reconstruction and Segmentation with a Unified Deep Network
May 06, 2018



Abstract:The need for fast acquisition and automatic analysis of MRI data is growing in the age of big data. Although compressed sensing magnetic resonance imaging (CS-MRI) has been studied to accelerate MRI by reducing k-space measurements, in current CS-MRI techniques MRI applications such as segmentation are overlooked when doing image reconstruction. In this paper, we test the utility of CS-MRI methods in automatic segmentation models and propose a unified deep neural network architecture called SegNetMRI which we apply to the combined CS-MRI reconstruction and segmentation problem. SegNetMRI is built upon a MRI reconstruction network with multiple cascaded blocks each containing an encoder-decoder unit and a data fidelity unit, and MRI segmentation networks having the same encoder-decoder structure. The two subnetworks are pre-trained and fine-tuned with shared reconstruction encoders. The outputs are merged into the final segmentation. Our experiments show that SegNetMRI can improve both the reconstruction and segmentation performance when using compressive measurements.
A Deep Information Sharing Network for Multi-contrast Compressed Sensing MRI Reconstruction
Apr 10, 2018



Abstract:In multi-contrast magnetic resonance imaging (MRI), compressed sensing theory can accelerate imaging by sampling fewer measurements within each contrast. The conventional optimization-based models suffer several limitations: strict assumption of shared sparse support, time-consuming optimization and "shallow" models with difficulties in encoding the rich patterns hiding in massive MRI data. In this paper, we propose the first deep learning model for multi-contrast MRI reconstruction. We achieve information sharing through feature sharing units, which significantly reduces the number of parameters. The feature sharing unit is combined with a data fidelity unit to comprise an inference block. These inference blocks are cascaded with dense connections, which allows for information transmission across different depths of the network efficiently. Our extensive experiments on various multi-contrast MRI datasets show that proposed model outperforms both state-of-the-art single-contrast and multi-contrast MRI methods in accuracy and efficiency. We show the improved reconstruction quality can bring great benefits for the later medical image analysis stage. Furthermore, the robustness of the proposed model to the non-registration environment shows its potential in real MRI applications.
A Segmentation-aware Deep Fusion Network for Compressed Sensing MRI
Apr 04, 2018



Abstract:Compressed sensing MRI is a classic inverse problem in the field of computational imaging, accelerating the MR imaging by measuring less k-space data. The deep neural network models provide the stronger representation ability and faster reconstruction compared with "shallow" optimization-based methods. However, in the existing deep-based CS-MRI models, the high-level semantic supervision information from massive segmentation-labels in MRI dataset is overlooked. In this paper, we proposed a segmentation-aware deep fusion network called SADFN for compressed sensing MRI. The multilayer feature aggregation (MLFA) method is introduced here to fuse all the features from different layers in the segmentation network. Then, the aggregated feature maps containing semantic information are provided to each layer in the reconstruction network with a feature fusion strategy. This guarantees the reconstruction network is aware of the different regions in the image it reconstructs, simplifying the function mapping. We prove the utility of the cross-layer and cross-task information fusion strategy by comparative study. Extensive experiments on brain segmentation benchmark MRBrainS validated that the proposed SADFN model achieves state-of-the-art accuracy in compressed sensing MRI. This paper provides a novel approach to guide the low-level visual task using the information from mid- or high-level task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge