Jiexiang Wang
LeTS: Learning to Think-and-Search via Process-and-Outcome Reward Hybridization
May 23, 2025Abstract:Large language models (LLMs) have demonstrated impressive capabilities in reasoning with the emergence of reasoning models like OpenAI-o1 and DeepSeek-R1. Recent research focuses on integrating reasoning capabilities into the realm of retrieval-augmented generation (RAG) via outcome-supervised reinforcement learning (RL) approaches, while the correctness of intermediate think-and-search steps is usually neglected. To address this issue, we design a process-level reward module to mitigate the unawareness of intermediate reasoning steps in outcome-level supervision without additional annotation. Grounded on this, we propose Learning to Think-and-Search (LeTS), a novel framework that hybridizes stepwise process reward and outcome-based reward to current RL methods for RAG. Extensive experiments demonstrate the generalization and inference efficiency of LeTS across various RAG benchmarks. In addition, these results reveal the potential of process- and outcome-level reward hybridization in boosting LLMs' reasoning ability via RL under other scenarios. The code will be released soon.
Multimodal Prompt Perceiver: Empower Adaptiveness, Generalizability and Fidelity for All-in-One Image Restoration
Dec 05, 2023Abstract:Despite substantial progress, all-in-one image restoration (IR) grapples with persistent challenges in handling intricate real-world degradations. This paper introduces MPerceiver: a novel multimodal prompt learning approach that harnesses Stable Diffusion (SD) priors to enhance adaptiveness, generalizability and fidelity for all-in-one image restoration. Specifically, we develop a dual-branch module to master two types of SD prompts: textual for holistic representation and visual for multiscale detail representation. Both prompts are dynamically adjusted by degradation predictions from the CLIP image encoder, enabling adaptive responses to diverse unknown degradations. Moreover, a plug-in detail refinement module improves restoration fidelity via direct encoder-to-decoder information transformation. To assess our method, MPerceiver is trained on 9 tasks for all-in-one IR and outperforms state-of-the-art task-specific methods across most tasks. Post multitask pre-training, MPerceiver attains a generalized representation in low-level vision, exhibiting remarkable zero-shot and few-shot capabilities in unseen tasks. Extensive experiments on 16 IR tasks and 26 benchmarks underscore the superiority of MPerceiver in terms of adaptiveness, generalizability and fidelity.
Unsupervised Adaptation of Polyp Segmentation Models via Coarse-to-Fine Self-Supervision
Aug 13, 2023Abstract:Unsupervised Domain Adaptation~(UDA) has attracted a surge of interest over the past decade but is difficult to be used in real-world applications. Considering the privacy-preservation issues and security concerns, in this work, we study a practical problem of Source-Free Domain Adaptation (SFDA), which eliminates the reliance on annotated source data. Current SFDA methods focus on extracting domain knowledge from the source-trained model but neglects the intrinsic structure of the target domain. Moreover, they typically utilize pseudo labels for self-training in the target domain, but suffer from the notorious error accumulation problem. To address these issues, we propose a new SFDA framework, called Region-to-Pixel Adaptation Network~(RPANet), which learns the region-level and pixel-level discriminative representations through coarse-to-fine self-supervision. The proposed RPANet consists of two modules, Foreground-aware Contrastive Learning (FCL) and Confidence-Calibrated Pseudo-Labeling (CCPL), which explicitly address the key challenges of ``how to distinguish'' and ``how to refine''. To be specific, FCL introduces a supervised contrastive learning paradigm in the region level to contrast different region centroids across different target images, which efficiently involves all pseudo labels while robust to noisy samples. CCPL designs a novel fusion strategy to reduce the overconfidence problem of pseudo labels by fusing two different target predictions without introducing any additional network modules. Extensive experiments on three cross-domain polyp segmentation tasks reveal that RPANet significantly outperforms state-of-the-art SFDA and UDA methods without access to source data, revealing the potential of SFDA in medical applications.
Digital Twin Brain: a simulation and assimilation platform for whole human brain
Aug 02, 2023Abstract:In this work, we present a computing platform named digital twin brain (DTB) that can simulate spiking neuronal networks of the whole human brain scale and more importantly, a personalized biological brain structure. In comparison to most brain simulations with a homogeneous global structure, we highlight that the sparseness, couplingness and heterogeneity in the sMRI, DTI and PET data of the brain has an essential impact on the efficiency of brain simulation, which is proved from the scaling experiments that the DTB of human brain simulation is communication-intensive and memory-access intensive computing systems rather than computation-intensive. We utilize a number of optimization techniques to balance and integrate the computation loads and communication traffics from the heterogeneous biological structure to the general GPU-based HPC and achieve leading simulation performance for the whole human brain-scaled spiking neuronal networks. On the other hand, the biological structure, equipped with a mesoscopic data assimilation, enables the DTB to investigate brain cognitive function by a reverse-engineering method, which is demonstrated by a digital experiment of visual evaluation on the DTB. Furthermore, we believe that the developing DTB will be a promising powerful platform for a large of research orients including brain-inspiredintelligence, rain disease medicine and brain-machine interface.
Multi-Task Neural Networks with Spatial Activation for Retinal Vessel Segmentation and Artery/Vein Classification
Jul 18, 2020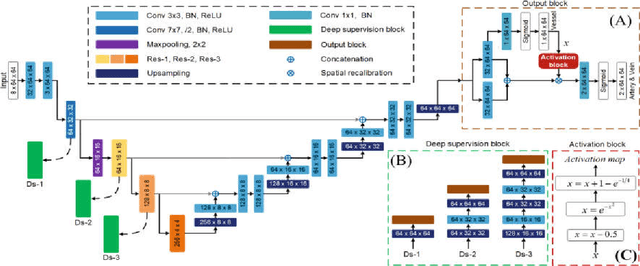
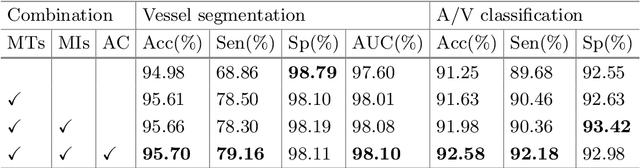
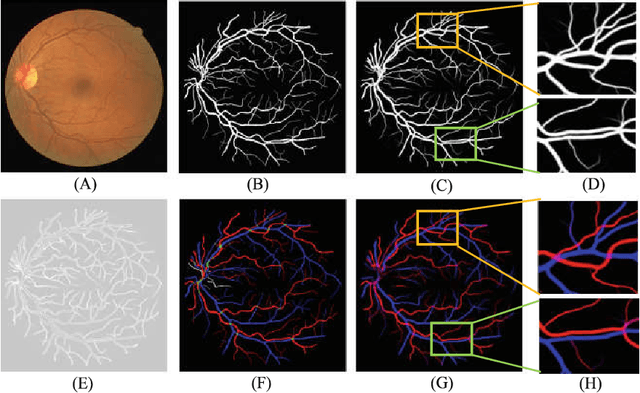
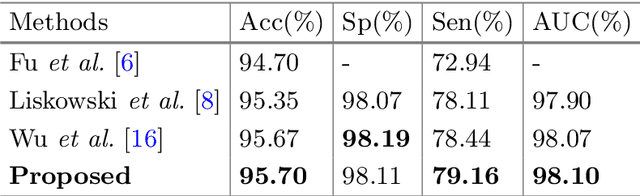
Abstract:Retinal artery/vein (A/V) classification plays a critical role in the clinical biomarker study of how various systemic and cardiovascular diseases affect the retinal vessels. Conventional methods of automated A/V classification are generally complicated and heavily depend on the accurate vessel segmentation. In this paper, we propose a multi-task deep neural network with spatial activation mechanism that is able to segment full retinal vessel, artery and vein simultaneously, without the pre-requirement of vessel segmentation. The input module of the network integrates the domain knowledge of widely used retinal preprocessing and vessel enhancement techniques. We specially customize the output block of the network with a spatial activation mechanism, which takes advantage of a relatively easier task of vessel segmentation and exploits it to boost the performance of A/V classification. In addition, deep supervision is introduced to the network to assist the low level layers to extract more semantic information. The proposed network achieves pixel-wise accuracy of 95.70% for vessel segmentation, and A/V classification accuracy of 94.50%, which is the state-of-the-art performance for both tasks on the AV-DRIVE dataset. Furthermore, we have also tested the model performance on INSPIRE-AVR dataset, which achieves a skeletal A/V classification accuracy of 91.6%.
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020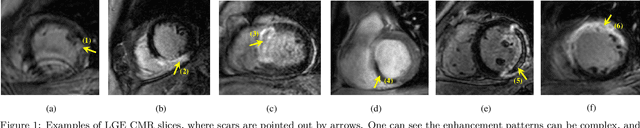
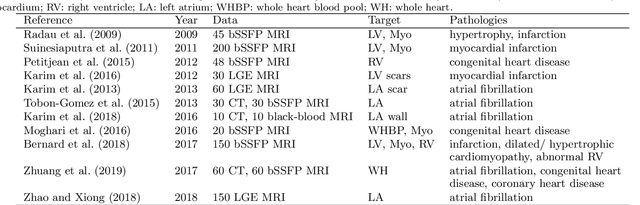
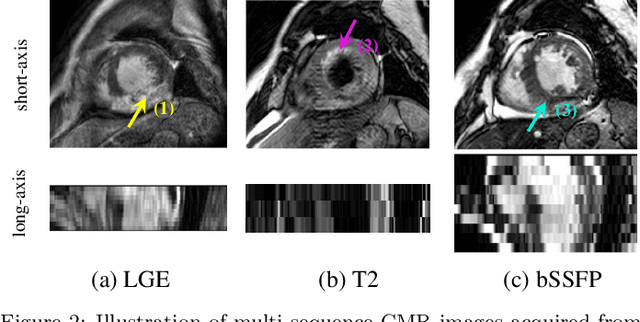
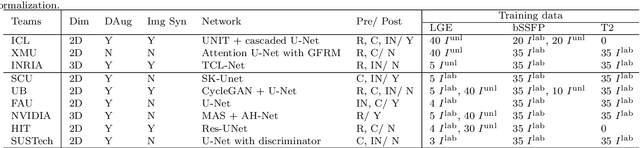
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
Multi-sequence Cardiac MR Segmentation with Adversarial Domain Adaptation Network
Oct 28, 2019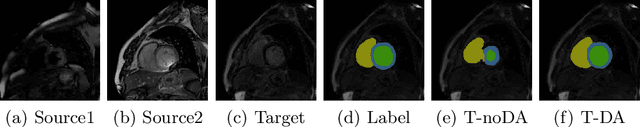
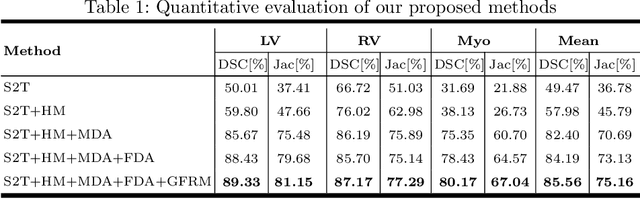
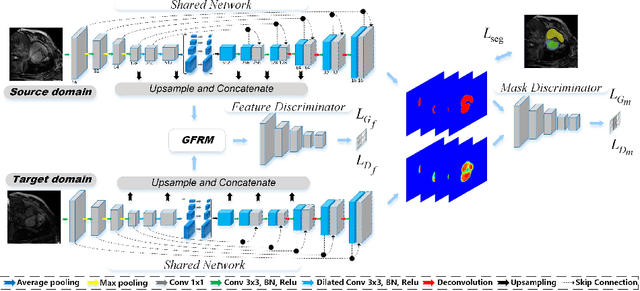
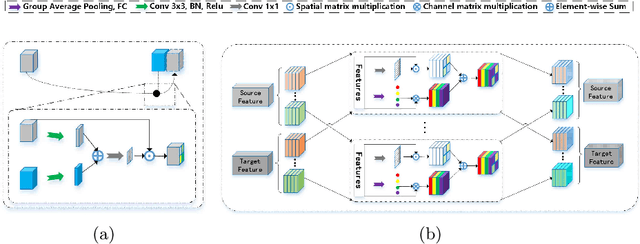
Abstract:Automatic and accurate segmentation of the ventricles and myocardium from multi-sequence cardiac MRI (CMR) is crucial for the diagnosis and treatment management for patients suffering from myocardial infarction (MI). However, due to the existence of domain shift among different modalities of datasets, the performance of deep neural networks drops significantly when the training and testing datasets are distinct. In this paper, we propose an unsupervised domain alignment method to explicitly alleviate the domain shifts among different modalities of CMR sequences, \emph{e.g.,} bSSFP, LGE, and T2-weighted. Our segmentation network is attention U-Net with pyramid pooling module, where multi-level feature space and output space adversarial learning are proposed to transfer discriminative domain knowledge across different datasets. Moreover, we further introduce a group-wise feature recalibration module to enforce the fine-grained semantic-level feature alignment that matching features from different networks but with the same class label. We evaluate our method on the multi-sequence cardiac MR Segmentation Challenge 2019 datasets, which contain three different modalities of MRI sequences. Extensive experimental results show that the proposed methods can obtain significant segmentation improvements compared with the baseline models.
Uncertainty-Guided Domain Alignment for Layer Segmentation in OCT Images
Aug 30, 2019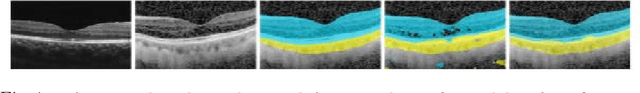
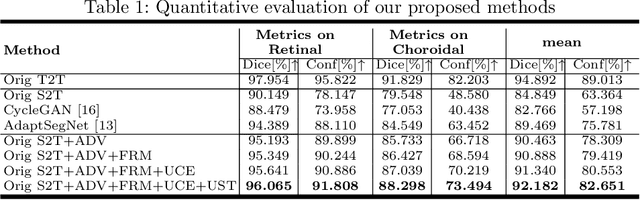
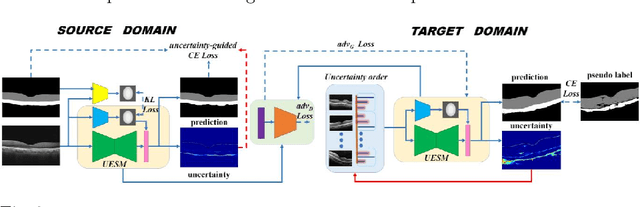
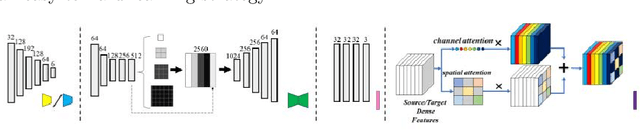
Abstract:Automatic and accurate segmentation for retinal and choroidal layers of Optical Coherence Tomography (OCT) is crucial for detection of various ocular diseases. However, because of the variations in different equipments, OCT data obtained from different manufacturers might encounter appearance discrepancy, which could lead to performance fluctuation to a deep neural network. In this paper, we propose an uncertainty-guided domain alignment method to aim at alleviating this problem to transfer discriminative knowledge across distinct domains. We disign a novel uncertainty-guided cross-entropy loss for boosting the performance over areas with high uncertainty. An uncertainty-guided curriculum transfer strategy is developed for the self-training (ST), which regards uncertainty as efficient and effective guidance to optimize the learning process in target domain. Adversarial learning with feature recalibration module (FRM) is applied to transfer informative knowledge from the domain feature spaces adaptively. The experiments on two OCT datasets show that the proposed methods can obtain significant segmentation improvements compared with the baseline models.
Distributed Cooperative Online Estimation With Random Observation Matrices, Communication Graphs and Time-Delays
Aug 22, 2019Abstract:We analyze convergence of distributed cooperative online estimation algorithms by a network of multiple nodes via information exchanging in an uncertain environment. Each node has a linear observation of an unknown parameter with randomly time-varying observation matrices. The underlying communication network is modeled by a sequence of random digraphs and is subjected to nonuniform random time-varying delays in channels. Each node runs an online estimation algorithm consisting of a consensus term taking a weighted sum of its own estimate and delayed estimates of neighbors, and an innovation term processing its own new measurement at each time step. By stochastic time-varying system, martingale convergence theories and the binomial expansion of random matrix products, we transform the convergence analysis of the algorithm into that of the mathematical expectation of random matrix products. Firstly, for the delay-free case, we show that the algorithm gains can be designed properly such that all nodes' estimates converge to the real parameter in mean square and almost surely if the observation matrices and communication graphs satisfy the stochastic spatial-temporal persistence of excitation condition. Especially, this condition holds for Markovian switching communication graphs and observation matrices, if the stationary graph is balanced with a spanning tree and the measurement model is spatially-temporally jointly observable. Secondly, for the case with time-delays, we introduce delay matrices to model the random time-varying communication delays between nodes, and propose a mean square convergence condition, which quantitatively shows the intensity of spatial-temporal persistence of excitation to overcome time-delays.
An Adversarial Learning Approach to Medical Image Synthesis for Lesion Removal
Oct 25, 2018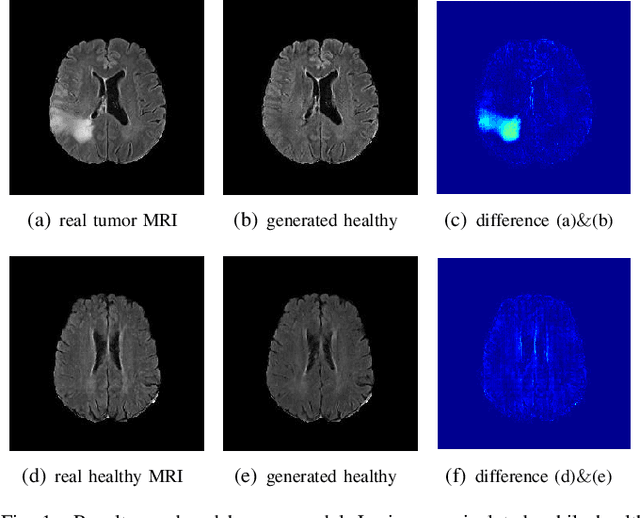
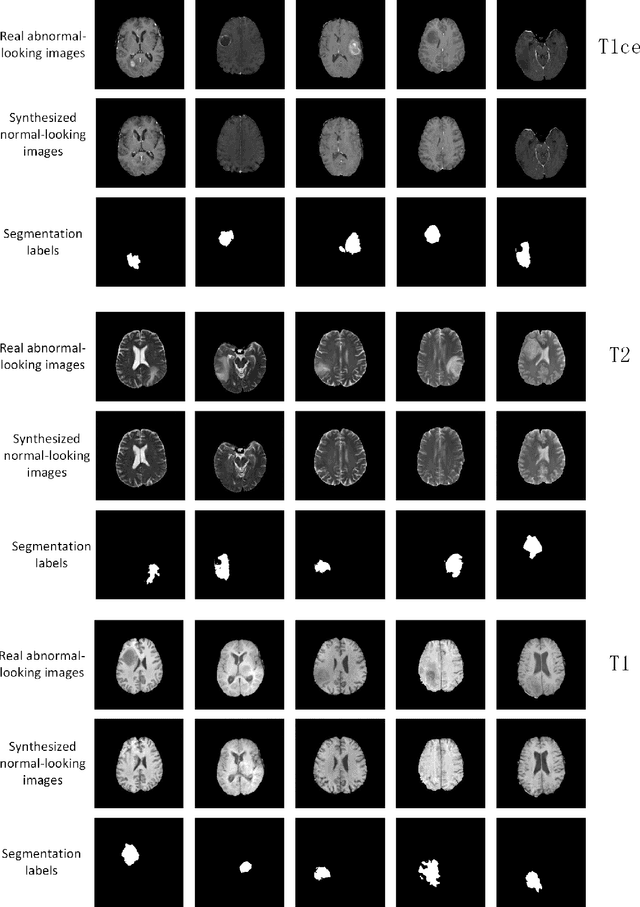
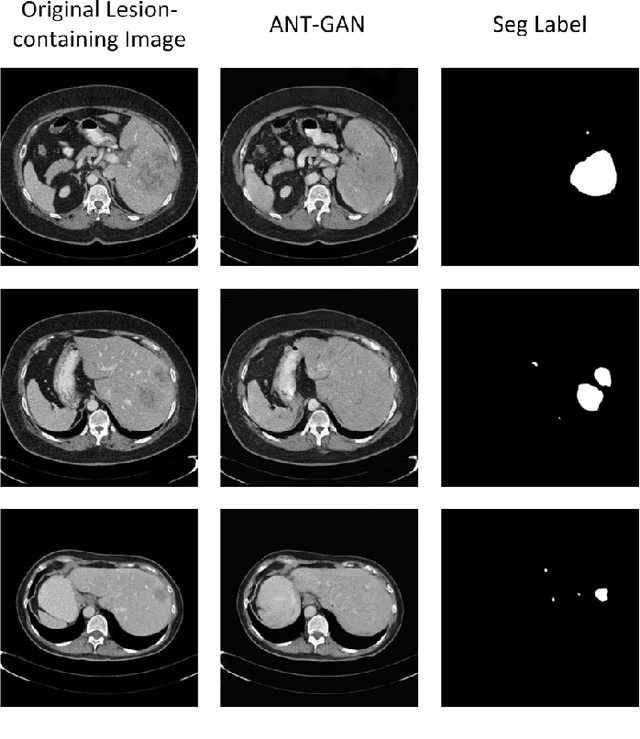
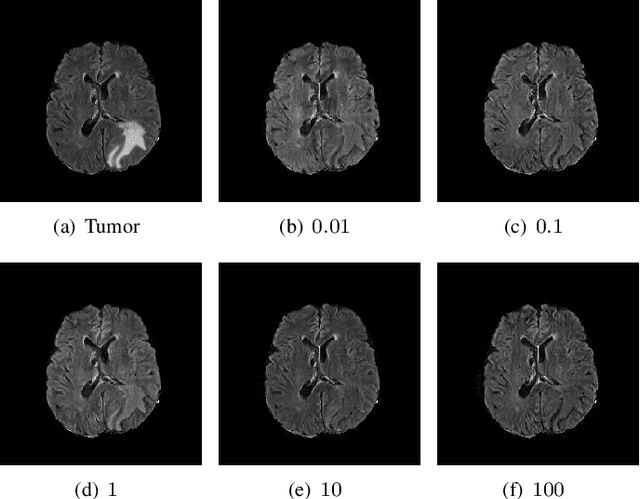
Abstract:The analysis of lesion within medical image data is desirable for efficient disease diagnosis, treatment and prognosis. The common lesion analysis tasks like segmentation and classification are mainly based on supervised learning with well-paired image-level or voxel-level labels. However, labeling the lesion in medical images is laborious requiring highly specialized knowledge. Inspired by the fact that radiologists make diagnoses based on expert knowledge on "healthiness" and "unhealthiness" developed from extensive experience, we propose an medical image synthesis model named abnormal-to-normal translation generative adversarial network (ANT-GAN) to predict a normal-looking medical image based on its abnormal-looking counterpart without the need of paired data for training. Unlike typical GANs, whose aim is to generate realistic samples with variations, our more restrictive model aims at producing the underlying normal-looking image corresponding to an image containing lesions, and thus requires a specialized design. With an ability to segment normal from abnormal tissue, our model is able to generate a highly realistic lesion-free medical image based on its true lesion-containing counterpart. Being able to provide a "normal" version of a medical image (possibly the same image if there is no illness) is not only an intriguing topic, but also can serve as a pre-processing and provide useful side information for medical imaging tasks like lesion segmentation or classification validated by our experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge