Dan Ruan
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025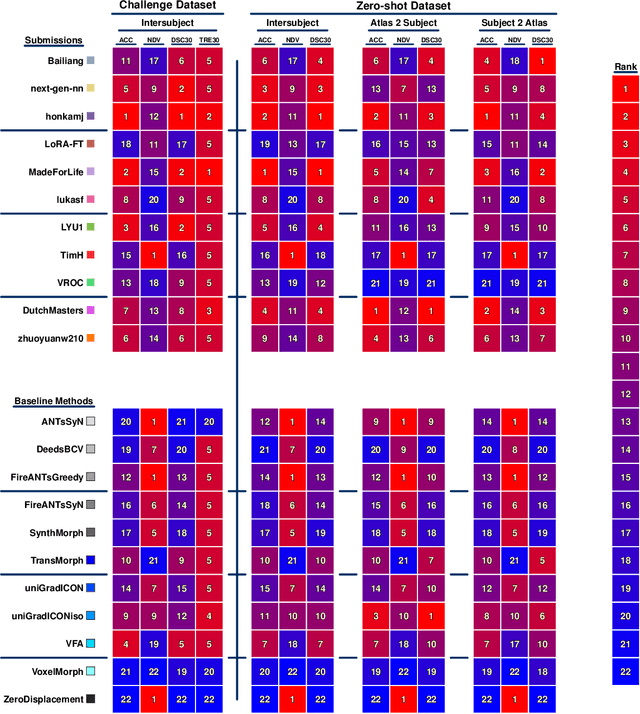
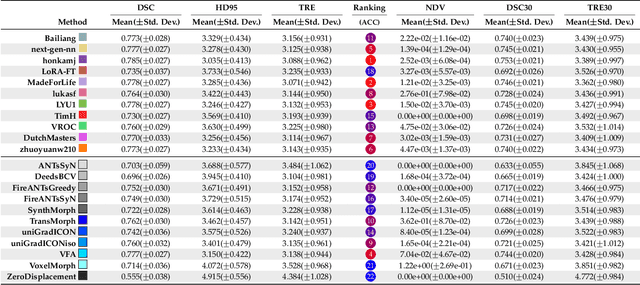
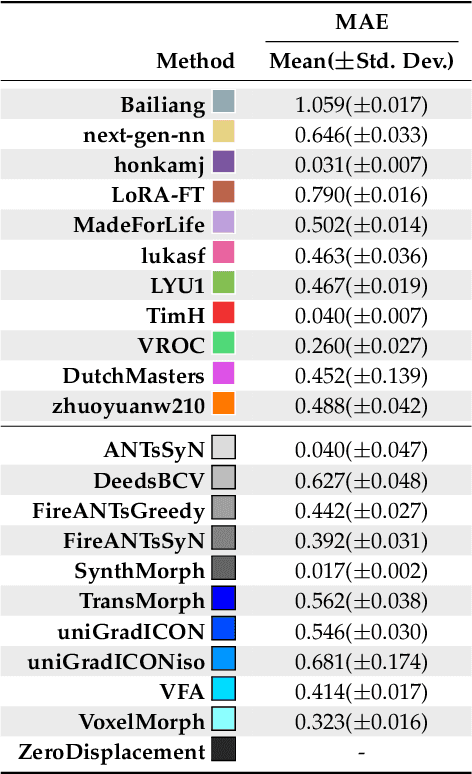
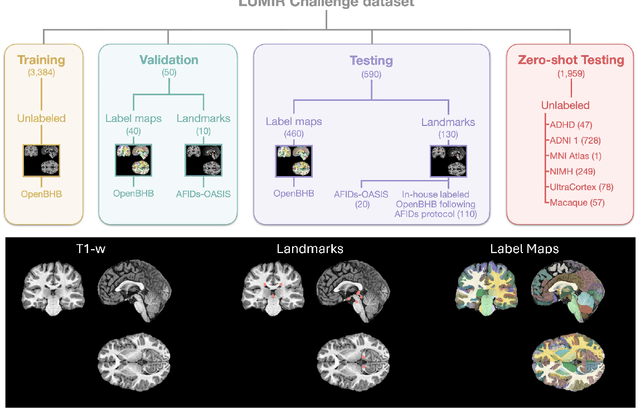
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Benchmark of Segmentation Techniques for Pelvic Fracture in CT and X-ray: Summary of the PENGWIN 2024 Challenge
Apr 03, 2025Abstract:The segmentation of pelvic fracture fragments in CT and X-ray images is crucial for trauma diagnosis, surgical planning, and intraoperative guidance. However, accurately and efficiently delineating the bone fragments remains a significant challenge due to complex anatomy and imaging limitations. The PENGWIN challenge, organized as a MICCAI 2024 satellite event, aimed to advance automated fracture segmentation by benchmarking state-of-the-art algorithms on these complex tasks. A diverse dataset of 150 CT scans was collected from multiple clinical centers, and a large set of simulated X-ray images was generated using the DeepDRR method. Final submissions from 16 teams worldwide were evaluated under a rigorous multi-metric testing scheme. The top-performing CT algorithm achieved an average fragment-wise intersection over union (IoU) of 0.930, demonstrating satisfactory accuracy. However, in the X-ray task, the best algorithm attained an IoU of 0.774, highlighting the greater challenges posed by overlapping anatomical structures. Beyond the quantitative evaluation, the challenge revealed methodological diversity in algorithm design. Variations in instance representation, such as primary-secondary classification versus boundary-core separation, led to differing segmentation strategies. Despite promising results, the challenge also exposed inherent uncertainties in fragment definition, particularly in cases of incomplete fractures. These findings suggest that interactive segmentation approaches, integrating human decision-making with task-relevant information, may be essential for improving model reliability and clinical applicability.
Accelerated Patient-specific Non-Cartesian MRI Reconstruction using Implicit Neural Representations
Mar 07, 2025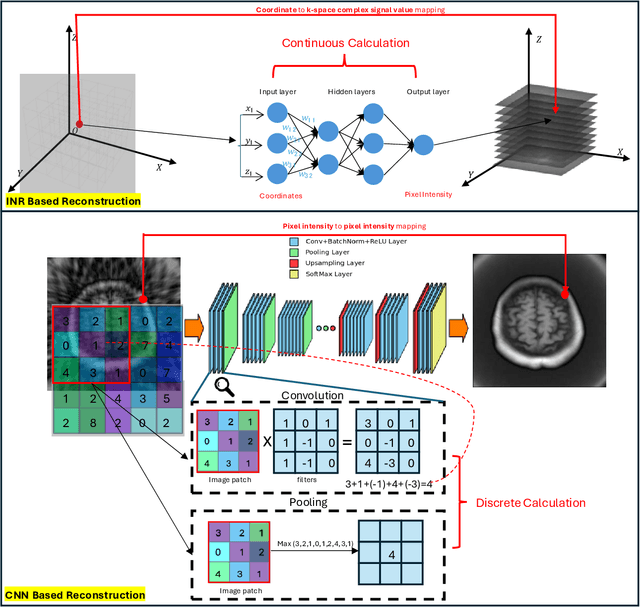
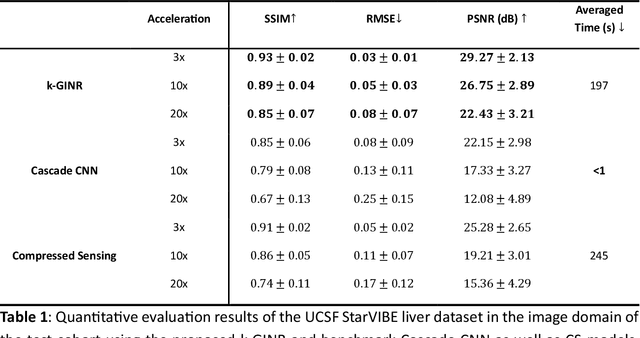
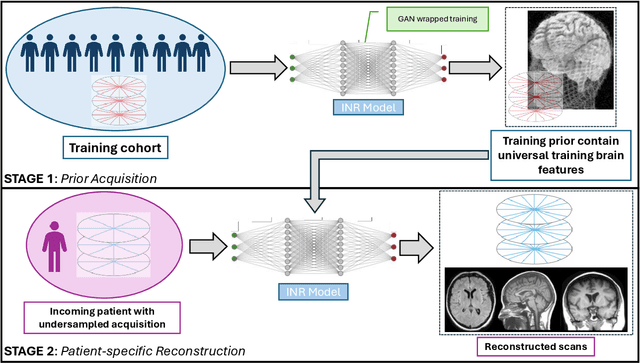
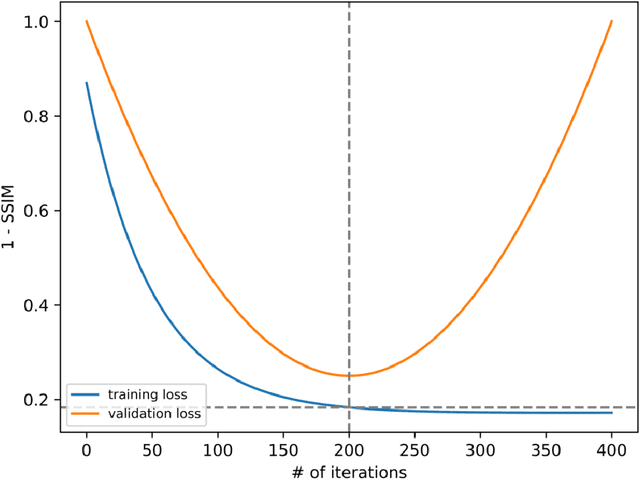
Abstract:The scanning time for a fully sampled MRI can be undesirably lengthy. Compressed sensing has been developed to minimize image artifacts in accelerated scans, but the required iterative reconstruction is computationally complex and difficult to generalize on new cases. Image-domain-based deep learning methods (e.g., convolutional neural networks) emerged as a faster alternative but face challenges in modeling continuous k-space, a problem amplified with non-Cartesian sampling commonly used in accelerated acquisition. In comparison, implicit neural representations can model continuous signals in the frequency domain and thus are compatible with arbitrary k-space sampling patterns. The current study develops a novel generative-adversarially trained implicit neural representations (k-GINR) for de novo undersampled non-Cartesian k-space reconstruction. k-GINR consists of two stages: 1) supervised training on an existing patient cohort; 2) self-supervised patient-specific optimization. In stage 1, the network is trained with the generative-adversarial network on diverse patients of the same anatomical region supervised by fully sampled acquisition. In stage 2, undersampled k-space data of individual patients is used to tailor the prior-embedded network for patient-specific optimization. The UCSF StarVIBE T1-weighted liver dataset was evaluated on the proposed framework. k-GINR is compared with an image-domain deep learning method, Deep Cascade CNN, and a compressed sensing method. k-GINR consistently outperformed the baselines with a larger performance advantage observed at very high accelerations (e.g., 20 times). k-GINR offers great value for direct non-Cartesian k-space reconstruction for new incoming patients across a wide range of accelerations liver anatomy.
TomoGRAF: A Robust and Generalizable Reconstruction Network for Single-View Computed Tomography
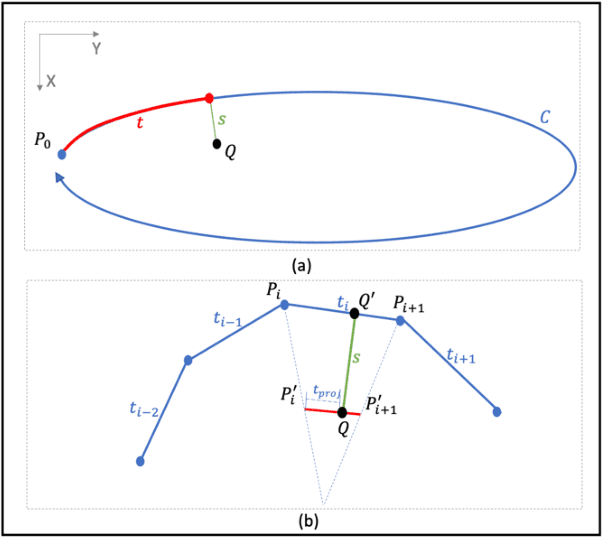
Nov 12, 2024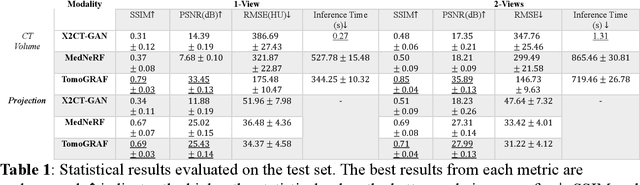

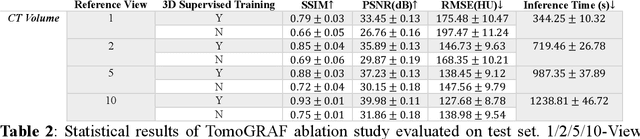

Abstract:Computed tomography (CT) provides high spatial resolution visualization of 3D structures for scientific and clinical applications. Traditional analytical/iterative CT reconstruction algorithms require hundreds of angular data samplings, a condition that may not be met in practice due to physical and mechanical limitations. Sparse view CT reconstruction has been proposed using constrained optimization and machine learning methods with varying success, less so for ultra-sparse view CT reconstruction with one to two views. Neural radiance field (NeRF) is a powerful tool for reconstructing and rendering 3D natural scenes from sparse views, but its direct application to 3D medical image reconstruction has been minimally successful due to the differences between optical and X-ray photon transportation. Here, we develop a novel TomoGRAF framework incorporating the unique X-ray transportation physics to reconstruct high-quality 3D volumes using ultra-sparse projections without prior. TomoGRAF captures the CT imaging geometry, simulates the X-ray casting and tracing process, and penalizes the difference between simulated and ground truth CT sub-volume during training. We evaluated the performance of TomoGRAF on an unseen dataset of distinct imaging characteristics from the training data and demonstrated a vast leap in performance compared with state-of-the-art deep learning and NeRF methods. TomoGRAF provides the first generalizable solution for image-guided radiotherapy and interventional radiology applications, where only one or a few X-ray views are available, but 3D volumetric information is desired.
Simultaneous Deep Learning of Myocardium Segmentation and T2 Quantification for Acute Myocardial Infarction MRI
May 17, 2024



Abstract:In cardiac Magnetic Resonance Imaging (MRI) analysis, simultaneous myocardial segmentation and T2 quantification are crucial for assessing myocardial pathologies. Existing methods often address these tasks separately, limiting their synergistic potential. To address this, we propose SQNet, a dual-task network integrating Transformer and Convolutional Neural Network (CNN) components. SQNet features a T2-refine fusion decoder for quantitative analysis, leveraging global features from the Transformer, and a segmentation decoder with multiple local region supervision for enhanced accuracy. A tight coupling module aligns and fuses CNN and Transformer branch features, enabling SQNet to focus on myocardium regions. Evaluation on healthy controls (HC) and acute myocardial infarction patients (AMI) demonstrates superior segmentation dice scores (89.3/89.2) compared to state-of-the-art methods (87.7/87.9). T2 quantification yields strong linear correlations (Pearson coefficients: 0.84/0.93) with label values for HC/AMI, indicating accurate mapping. Radiologist evaluations confirm SQNet's superior image quality scores (4.60/4.58 for segmentation, 4.32/4.42 for T2 quantification) over state-of-the-art methods (4.50/4.44 for segmentation, 3.59/4.37 for T2 quantification). SQNet thus offers accurate simultaneous segmentation and quantification, enhancing cardiac disease diagnosis, such as AMI.
A Two-Step Framework for Multi-Material Decomposition of Dual Energy Computed Tomography from Projection Domain
Oct 31, 2023Abstract:Dual-energy computed tomography (DECT) utilizes separate X-ray energy spectra to improve multi-material decomposition (MMD) for various diagnostic applications. However accurate decomposing more than two types of material remains challenging using conventional methods. Deep learning (DL) methods have shown promise to improve the MMD performance, but typical approaches of conducing DL-MMD in the image domain fail to fully utilize projection information or under iterative setup are computationally inefficient in both training and prediction. In this work, we present a clinical-applicable MMD (>2) framework rFast-MMDNet, operating with raw projection data in non-recursive setup, for breast tissue differentiation. rFast-MMDNet is a two-stage algorithm, including stage-one SinoNet to perform dual energy projection decomposition on tissue sinograms and stage-two FBP-DenoiseNet to perform domain adaptation and image post-processing. rFast-MMDNet was tested on a 2022 DL-Spectral-Challenge breast phantom dataset. The two stages of rFast-MMDNet were evaluated separately and then compared with four noniterative reference methods including a direct inversion method (AA-MMD), an image domain DL method (ID-UNet), AA-MMD/ID-UNet + DenoiseNet and a sinogram domain DL method (Triple-CBCT). Our results show that models trained from information stored in DE transmission domain can yield high-fidelity decomposition of the adipose, calcification, and fibroglandular materials with averaged RMSE, MAE, negative PSNR, and SSIM of 0.004+/-~0, 0.001+/-~0, -45.027+/-~0.542, and 0.002+/-~0 benchmarking to the ground truth, respectively. Training of entire rFast-MMDNet on a 4xRTX A6000 GPU cluster took a day with inference time <1s. All DL methods generally led to more accurate MMD than AA-MMD. rFast-MMDNet outperformed Triple-CBCT, but both are superior to the image-domain based methods.
Learning Dynamic MRI Reconstruction with Convolutional Network Assisted Reconstruction Swin Transformer
Sep 19, 2023
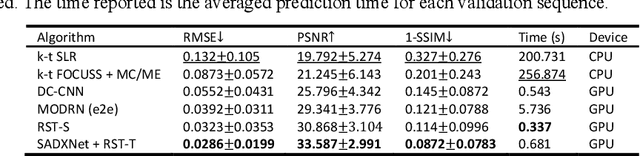

Abstract:Dynamic magnetic resonance imaging (DMRI) is an effective imaging tool for diagnosis tasks that require motion tracking of a certain anatomy. To speed up DMRI acquisition, k-space measurements are commonly undersampled along spatial or spatial-temporal domains. The difficulty of recovering useful information increases with increasing undersampling ratios. Compress sensing was invented for this purpose and has become the most popular method until deep learning (DL) based DMRI reconstruction methods emerged in the past decade. Nevertheless, existing DL networks are still limited in long-range sequential dependency understanding and computational efficiency and are not fully automated. Considering the success of Transformers positional embedding and "swin window" self-attention mechanism in the vision community, especially natural video understanding, we hereby propose a novel architecture named Reconstruction Swin Transformer (RST) for 4D MRI. RST inherits the backbone design of the Video Swin Transformer with a novel reconstruction head introduced to restore pixel-wise intensity. A convolution network called SADXNet is used for rapid initialization of 2D MR frames before RST learning to effectively reduce the model complexity, GPU hardware demand, and training time. Experimental results in the cardiac 4D MR dataset further substantiate the superiority of RST, achieving the lowest RMSE of 0.0286 +/- 0.0199 and 1 - SSIM of 0.0872 +/- 0.0783 on 9 times accelerated validation sequences.
An Efficient and Robust Method for Chest X-Ray Rib Suppression that Improves Pulmonary Abnormality Diagnosis
Feb 19, 2023


Abstract:Suppression of thoracic bone shadows on chest X-rays (CXRs) has been indicated to improve the diagnosis of pulmonary disease. Previous approaches can be categorized as unsupervised physical and supervised deep learning models. Nevertheless, with physical models able to preserve morphological details but at the cost of extremely long processing time, existing DL methods face challenges of gathering sufficient/qualitative ground truth (GT) for robust training, thus leading to failure in maintaining clinically acceptable false positive rates. We hereby propose a generalizable yet efficient workflow of two stages: (1) training pairs generation with GT bone shadows eliminated in by a physical model in spatially transformed gradient fields. (2) fully supervised image denoising network training on stage-one datasets for fast rib removal on incoming CXRs. For step two, we designed a densely connected network called SADXNet, combined with peak signal to noise ratio and multi-scale structure similarity index measure objective minimization to suppress bony structures. The SADXNet organizes spatial filters in U shape (e.g., X=7; filters = 16, 64, 256, 512, 256, 64, 16) and preserves the feature map dimension throughout the network flow. Visually, SADXNet can suppress the rib edge and that near the lung wall/vertebra without jeopardizing the vessel/abnormality conspicuity. Quantitively, it achieves RMSE of ~0 during testing with one prediction taking <1s. Downstream tasks including lung nodule detection as well as common lung disease classification and localization are used to evaluate our proposed rib suppression mechanism. We observed 3.23% and 6.62% area under the curve (AUC) increase as well as 203 and 385 absolute false positive decrease for lung nodule detection and common lung disease localization, separately.
To what extent can Plug-and-Play methods outperform neural networks alone in low-dose CT reconstruction
Feb 15, 2022
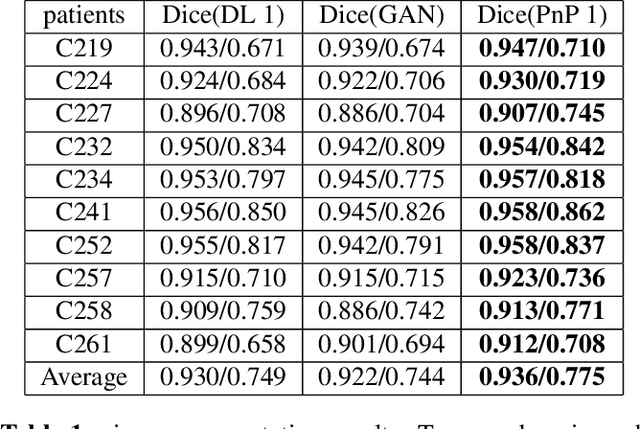


Abstract:The Plug-and-Play (PnP) framework was recently introduced for low-dose CT reconstruction to leverage the interpretability and the flexibility of model-based methods to incorporate various plugins, such as trained deep learning (DL) neural networks. However, the benefits of PnP vs. state-of-the-art DL methods have not been clearly demonstrated. In this work, we proposed an improved PnP framework to address the previous limitations and develop clinical-relevant segmentation metrics for quantitative result assessment. Compared with the DL alone methods, our proposed PnP framework was slightly inferior in MSE and PSNR. However, the power spectrum of the resulting images better matched that of full-dose images than that of DL denoised images. The resulting images supported higher accuracy in airway segmentation than DL denoised images for all the ten patients in the test set, more substantially on the airways with a cross-section smaller than 0.61cm$^2$, and outperformed the DL denoised images for 45 out of 50 lung lobes in lobar segmentation. Our PnP method proved to be significantly better at preserving the image texture, which translated to task-specific benefits in automated structure segmentation and detection.
Fully Automated Multi-Organ Segmentation in Abdominal Magnetic Resonance Imaging with Deep Neural Networks
Dec 23, 2019



Abstract:Segmentation of multiple organs-at-risk (OARs) is essential for radiation therapy treatment planning and other clinical applications. We developed an Automated deep Learning-based Abdominal Multi-Organ segmentation (ALAMO) framework based on 2D U-net and a densely connected network structure with tailored design in data augmentation and training procedures such as deep connection, auxiliary supervision, and multi-view. The model takes in multi-slice MR images and generates the output of segmentation results. Three-Tesla T1 VIBE (Volumetric Interpolated Breath-hold Examination) images of 102 subjects were collected and used in our study. Ten OARs were studied, including the liver, spleen, pancreas, left/right kidneys, stomach, duodenum, small intestine, spinal cord, and vertebral bodies. Two radiologists manually labeled and obtained the consensus contours as the ground-truth. In the complete cohort of 102, 20 samples were held out for independent testing, and the rest were used for training and validation. The performance was measured using volume overlapping and surface distance. The ALAMO framework generated segmentation labels in good agreement with the manual results. Specifically, among the 10 OARs, 9 achieved high Dice Similarity Coefficients (DSCs) in the range of 0.87-0.96, except for the duodenum with a DSC of 0.80. The inference completes within one minute for a 3D volume of 320x288x180. Overall, the ALAMO model matches the state-of-the-art performance. The proposed ALAMO framework allows for fully automated abdominal MR segmentation with high accuracy and low memory and computation time demands.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge