Michael Ohliger
Accelerated Patient-specific Non-Cartesian MRI Reconstruction using Implicit Neural Representations
Mar 07, 2025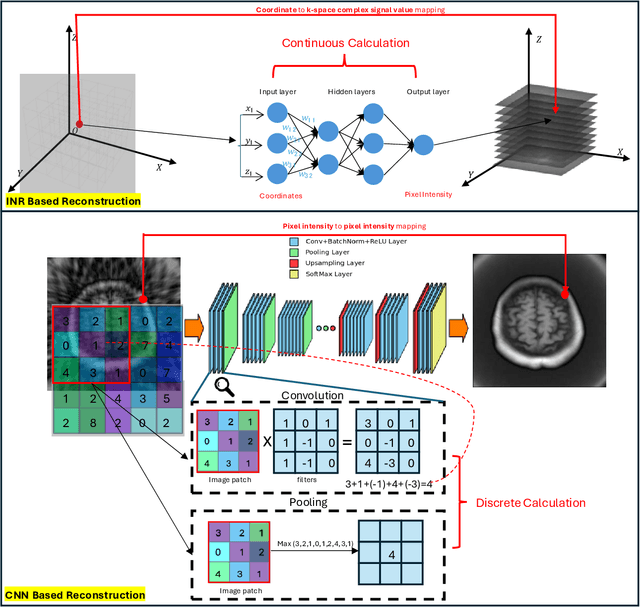
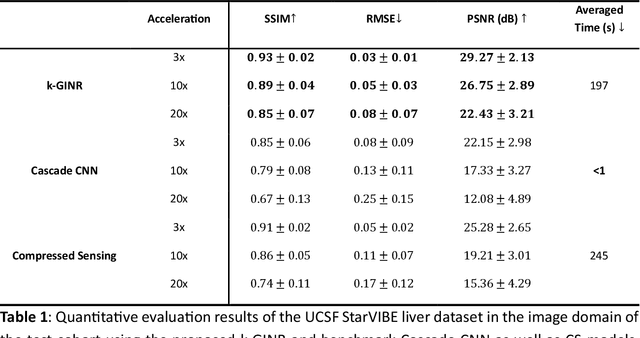
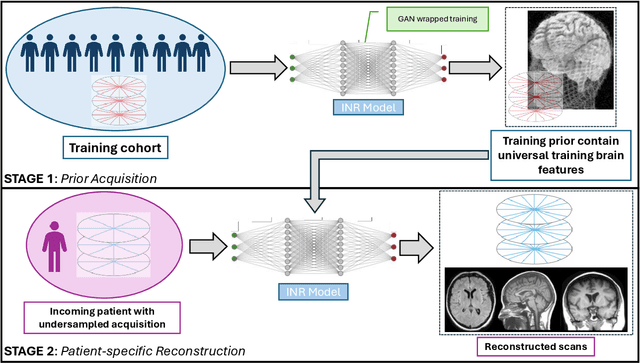
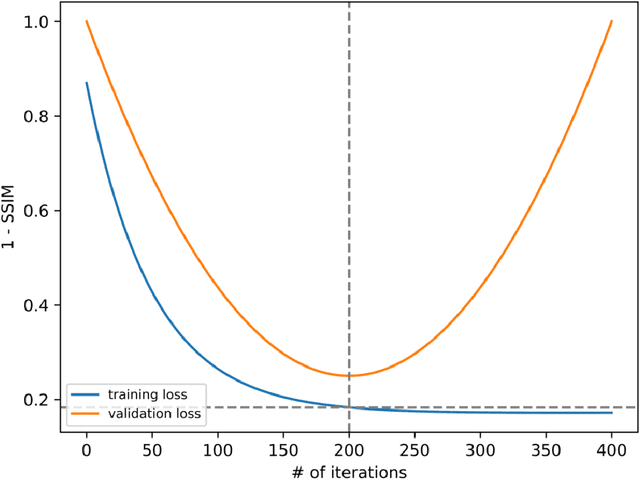
Abstract:The scanning time for a fully sampled MRI can be undesirably lengthy. Compressed sensing has been developed to minimize image artifacts in accelerated scans, but the required iterative reconstruction is computationally complex and difficult to generalize on new cases. Image-domain-based deep learning methods (e.g., convolutional neural networks) emerged as a faster alternative but face challenges in modeling continuous k-space, a problem amplified with non-Cartesian sampling commonly used in accelerated acquisition. In comparison, implicit neural representations can model continuous signals in the frequency domain and thus are compatible with arbitrary k-space sampling patterns. The current study develops a novel generative-adversarially trained implicit neural representations (k-GINR) for de novo undersampled non-Cartesian k-space reconstruction. k-GINR consists of two stages: 1) supervised training on an existing patient cohort; 2) self-supervised patient-specific optimization. In stage 1, the network is trained with the generative-adversarial network on diverse patients of the same anatomical region supervised by fully sampled acquisition. In stage 2, undersampled k-space data of individual patients is used to tailor the prior-embedded network for patient-specific optimization. The UCSF StarVIBE T1-weighted liver dataset was evaluated on the proposed framework. k-GINR is compared with an image-domain deep learning method, Deep Cascade CNN, and a compressed sensing method. k-GINR consistently outperformed the baselines with a larger performance advantage observed at very high accelerations (e.g., 20 times). k-GINR offers great value for direct non-Cartesian k-space reconstruction for new incoming patients across a wide range of accelerations liver anatomy.
Rapid Reconstruction of Extremely Accelerated Liver 4D MRI via Chained Iterative Refinement
Dec 14, 2024



Abstract:Abstract Purpose: High-quality 4D MRI requires an impractically long scanning time for dense k-space signal acquisition covering all respiratory phases. Accelerated sparse sampling followed by reconstruction enhancement is desired but often results in degraded image quality and long reconstruction time. We hereby propose the chained iterative reconstruction network (CIRNet) for efficient sparse-sampling reconstruction while maintaining clinically deployable quality. Methods: CIRNet adopts the denoising diffusion probabilistic framework to condition the image reconstruction through a stochastic iterative denoising process. During training, a forward Markovian diffusion process is designed to gradually add Gaussian noise to the densely sampled ground truth (GT), while CIRNet is optimized to iteratively reverse the Markovian process from the forward outputs. At the inference stage, CIRNet performs the reverse process solely to recover signals from noise, conditioned upon the undersampled input. CIRNet processed the 4D data (3D+t) as temporal slices (2D+t). The proposed framework is evaluated on a data cohort consisting of 48 patients (12332 temporal slices) who underwent free-breathing liver 4D MRI. 3-, 6-, 10-, 20- and 30-times acceleration were examined with a retrospective random undersampling scheme. Compressed sensing (CS) reconstruction with a spatiotemporal constraint and a recently proposed deep network, Re-Con-GAN, are selected as baselines. Results: CIRNet consistently achieved superior performance compared to CS and Re-Con-GAN. The inference time of CIRNet, CS, and Re-Con-GAN are 11s, 120s, and 0.15s. Conclusion: A novel framework, CIRNet, is presented. CIRNet maintains useable image quality for acceleration up to 30 times, significantly reducing the burden of 4DMRI.
Paired Conditional Generative Adversarial Network for Highly Accelerated Liver 4D MRI
May 20, 2024Abstract:Purpose: 4D MRI with high spatiotemporal resolution is desired for image-guided liver radiotherapy. Acquiring densely sampling k-space data is time-consuming. Accelerated acquisition with sparse samples is desirable but often causes degraded image quality or long reconstruction time. We propose the Reconstruct Paired Conditional Generative Adversarial Network (Re-Con-GAN) to shorten the 4D MRI reconstruction time while maintaining the reconstruction quality. Methods: Patients who underwent free-breathing liver 4D MRI were included in the study. Fully- and retrospectively under-sampled data at 3, 6 and 10 times (3x, 6x and 10x) were first reconstructed using the nuFFT algorithm. Re-Con-GAN then trained input and output in pairs. Three types of networks, ResNet9, UNet and reconstruction swin transformer, were explored as generators. PatchGAN was selected as the discriminator. Re-Con-GAN processed the data (3D+t) as temporal slices (2D+t). A total of 48 patients with 12332 temporal slices were split into training (37 patients with 10721 slices) and test (11 patients with 1611 slices). Results: Re-Con-GAN consistently achieved comparable/better PSNR, SSIM, and RMSE scores compared to CS/UNet models. The inference time of Re-Con-GAN, UNet and CS are 0.15s, 0.16s, and 120s. The GTV detection task showed that Re-Con-GAN and CS, compared to UNet, better improved the dice score (3x Re-Con-GAN 80.98%; 3x CS 80.74%; 3x UNet 79.88%) of unprocessed under-sampled images (3x 69.61%). Conclusion: A generative network with adversarial training is proposed with promising and efficient reconstruction results demonstrated on an in-house dataset. The rapid and qualitative reconstruction of 4D liver MR has the potential to facilitate online adaptive MR-guided radiotherapy for liver cancer.
Advancing low-field MRI with a universal denoising imaging transformer: Towards fast and high-quality imaging
Apr 30, 2024Abstract:Recent developments in low-field (LF) magnetic resonance imaging (MRI) systems present remarkable opportunities for affordable and widespread MRI access. A robust denoising method to overcome the intrinsic low signal-noise-ratio (SNR) barrier is critical to the success of LF MRI. However, current data-driven MRI denoising methods predominantly handle magnitude images and rely on customized models with constrained data diversity and quantity, which exhibit limited generalizability in clinical applications across diverse MRI systems, pulse sequences, and organs. In this study, we present ImT-MRD: a complex-valued imaging transformer trained on a vast number of clinical MRI scans aiming at universal MR denoising at LF systems. Compared with averaging multiple-repeated scans for higher image SNR, the model obtains better image quality from fewer repetitions, demonstrating its capability for accelerating scans under various clinical settings. Moreover, with its complex-valued image input, the model can denoise intermediate results before advanced post-processing and prepare high-quality data for further MRI research. By delivering universal and accurate denoising across clinical and research tasks, our model holds great promise to expedite the evolution of LF MRI for accessible and equal biomedical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge