Hengjie Liu
Accelerated Patient-specific Non-Cartesian MRI Reconstruction using Implicit Neural Representations
Mar 07, 2025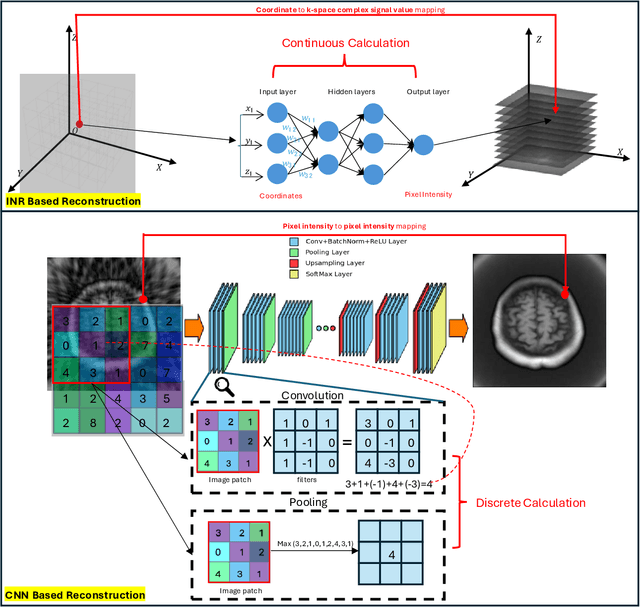
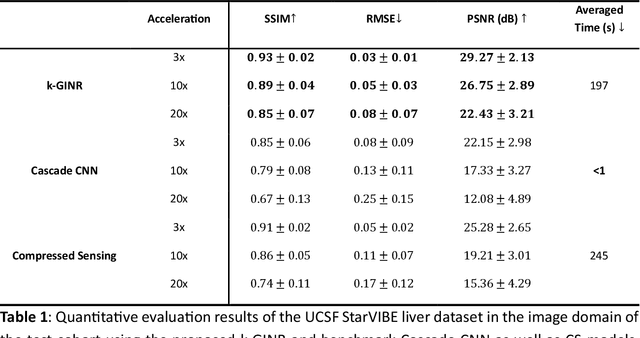
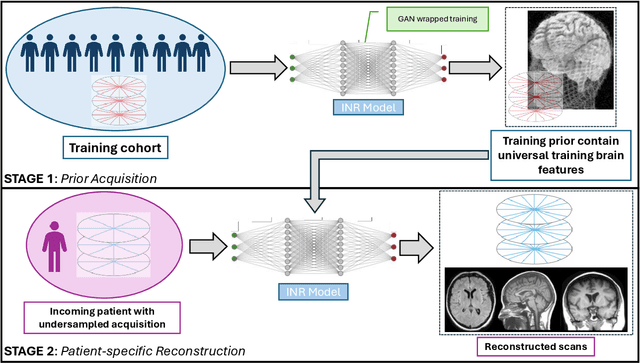
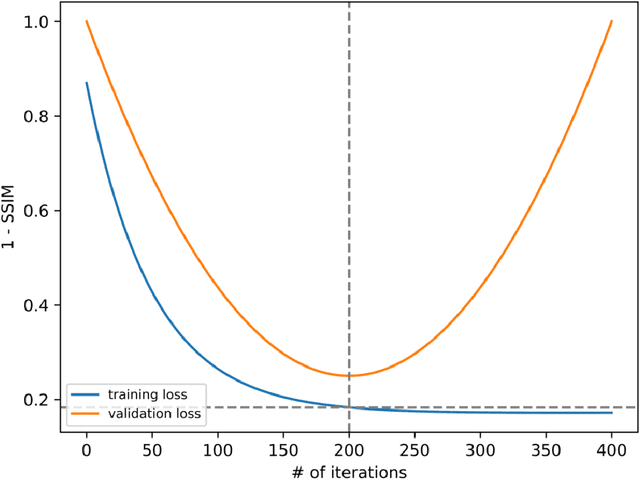
Abstract:The scanning time for a fully sampled MRI can be undesirably lengthy. Compressed sensing has been developed to minimize image artifacts in accelerated scans, but the required iterative reconstruction is computationally complex and difficult to generalize on new cases. Image-domain-based deep learning methods (e.g., convolutional neural networks) emerged as a faster alternative but face challenges in modeling continuous k-space, a problem amplified with non-Cartesian sampling commonly used in accelerated acquisition. In comparison, implicit neural representations can model continuous signals in the frequency domain and thus are compatible with arbitrary k-space sampling patterns. The current study develops a novel generative-adversarially trained implicit neural representations (k-GINR) for de novo undersampled non-Cartesian k-space reconstruction. k-GINR consists of two stages: 1) supervised training on an existing patient cohort; 2) self-supervised patient-specific optimization. In stage 1, the network is trained with the generative-adversarial network on diverse patients of the same anatomical region supervised by fully sampled acquisition. In stage 2, undersampled k-space data of individual patients is used to tailor the prior-embedded network for patient-specific optimization. The UCSF StarVIBE T1-weighted liver dataset was evaluated on the proposed framework. k-GINR is compared with an image-domain deep learning method, Deep Cascade CNN, and a compressed sensing method. k-GINR consistently outperformed the baselines with a larger performance advantage observed at very high accelerations (e.g., 20 times). k-GINR offers great value for direct non-Cartesian k-space reconstruction for new incoming patients across a wide range of accelerations liver anatomy.
Rapid Reconstruction of Extremely Accelerated Liver 4D MRI via Chained Iterative Refinement
Dec 14, 2024



Abstract:Abstract Purpose: High-quality 4D MRI requires an impractically long scanning time for dense k-space signal acquisition covering all respiratory phases. Accelerated sparse sampling followed by reconstruction enhancement is desired but often results in degraded image quality and long reconstruction time. We hereby propose the chained iterative reconstruction network (CIRNet) for efficient sparse-sampling reconstruction while maintaining clinically deployable quality. Methods: CIRNet adopts the denoising diffusion probabilistic framework to condition the image reconstruction through a stochastic iterative denoising process. During training, a forward Markovian diffusion process is designed to gradually add Gaussian noise to the densely sampled ground truth (GT), while CIRNet is optimized to iteratively reverse the Markovian process from the forward outputs. At the inference stage, CIRNet performs the reverse process solely to recover signals from noise, conditioned upon the undersampled input. CIRNet processed the 4D data (3D+t) as temporal slices (2D+t). The proposed framework is evaluated on a data cohort consisting of 48 patients (12332 temporal slices) who underwent free-breathing liver 4D MRI. 3-, 6-, 10-, 20- and 30-times acceleration were examined with a retrospective random undersampling scheme. Compressed sensing (CS) reconstruction with a spatiotemporal constraint and a recently proposed deep network, Re-Con-GAN, are selected as baselines. Results: CIRNet consistently achieved superior performance compared to CS and Re-Con-GAN. The inference time of CIRNet, CS, and Re-Con-GAN are 11s, 120s, and 0.15s. Conclusion: A novel framework, CIRNet, is presented. CIRNet maintains useable image quality for acceleration up to 30 times, significantly reducing the burden of 4DMRI.
TomoGRAF: A Robust and Generalizable Reconstruction Network for Single-View Computed Tomography
Nov 12, 2024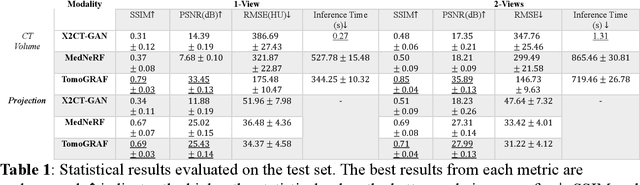

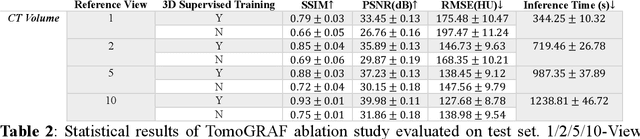

Abstract:Computed tomography (CT) provides high spatial resolution visualization of 3D structures for scientific and clinical applications. Traditional analytical/iterative CT reconstruction algorithms require hundreds of angular data samplings, a condition that may not be met in practice due to physical and mechanical limitations. Sparse view CT reconstruction has been proposed using constrained optimization and machine learning methods with varying success, less so for ultra-sparse view CT reconstruction with one to two views. Neural radiance field (NeRF) is a powerful tool for reconstructing and rendering 3D natural scenes from sparse views, but its direct application to 3D medical image reconstruction has been minimally successful due to the differences between optical and X-ray photon transportation. Here, we develop a novel TomoGRAF framework incorporating the unique X-ray transportation physics to reconstruct high-quality 3D volumes using ultra-sparse projections without prior. TomoGRAF captures the CT imaging geometry, simulates the X-ray casting and tracing process, and penalizes the difference between simulated and ground truth CT sub-volume during training. We evaluated the performance of TomoGRAF on an unseen dataset of distinct imaging characteristics from the training data and demonstrated a vast leap in performance compared with state-of-the-art deep learning and NeRF methods. TomoGRAF provides the first generalizable solution for image-guided radiotherapy and interventional radiology applications, where only one or a few X-ray views are available, but 3D volumetric information is desired.
High-Quality Data Augmentation for Low-Resource NMT: Combining a Translation Memory, a GAN Generator, and Filtering
Aug 22, 2024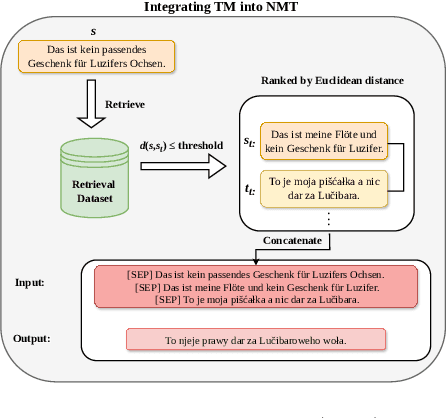
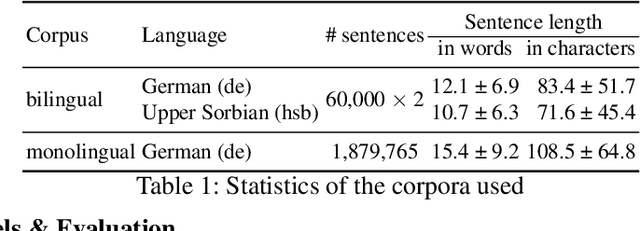
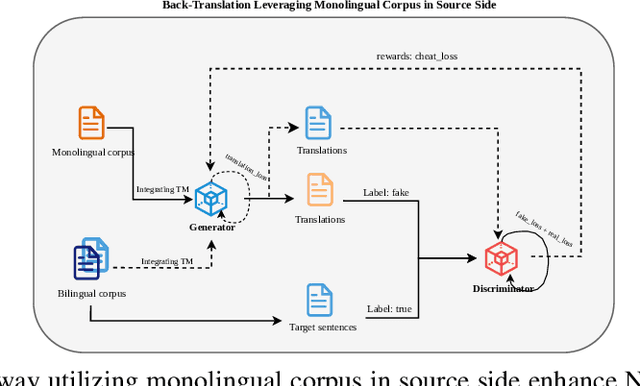
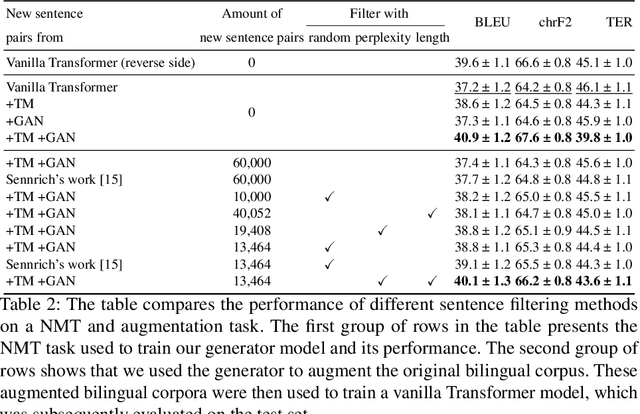
Abstract:Back translation, as a technique for extending a dataset, is widely used by researchers in low-resource language translation tasks. It typically translates from the target to the source language to ensure high-quality translation results. This paper proposes a novel way of utilizing a monolingual corpus on the source side to assist Neural Machine Translation (NMT) in low-resource settings. We realize this concept by employing a Generative Adversarial Network (GAN), which augments the training data for the discriminator while mitigating the interference of low-quality synthetic monolingual translations with the generator. Additionally, this paper integrates Translation Memory (TM) with NMT, increasing the amount of data available to the generator. Moreover, we propose a novel procedure to filter the synthetic sentence pairs during the augmentation process, ensuring the high quality of the data.
Paired Conditional Generative Adversarial Network for Highly Accelerated Liver 4D MRI
May 20, 2024Abstract:Purpose: 4D MRI with high spatiotemporal resolution is desired for image-guided liver radiotherapy. Acquiring densely sampling k-space data is time-consuming. Accelerated acquisition with sparse samples is desirable but often causes degraded image quality or long reconstruction time. We propose the Reconstruct Paired Conditional Generative Adversarial Network (Re-Con-GAN) to shorten the 4D MRI reconstruction time while maintaining the reconstruction quality. Methods: Patients who underwent free-breathing liver 4D MRI were included in the study. Fully- and retrospectively under-sampled data at 3, 6 and 10 times (3x, 6x and 10x) were first reconstructed using the nuFFT algorithm. Re-Con-GAN then trained input and output in pairs. Three types of networks, ResNet9, UNet and reconstruction swin transformer, were explored as generators. PatchGAN was selected as the discriminator. Re-Con-GAN processed the data (3D+t) as temporal slices (2D+t). A total of 48 patients with 12332 temporal slices were split into training (37 patients with 10721 slices) and test (11 patients with 1611 slices). Results: Re-Con-GAN consistently achieved comparable/better PSNR, SSIM, and RMSE scores compared to CS/UNet models. The inference time of Re-Con-GAN, UNet and CS are 0.15s, 0.16s, and 120s. The GTV detection task showed that Re-Con-GAN and CS, compared to UNet, better improved the dice score (3x Re-Con-GAN 80.98%; 3x CS 80.74%; 3x UNet 79.88%) of unprocessed under-sampled images (3x 69.61%). Conclusion: A generative network with adversarial training is proposed with promising and efficient reconstruction results demonstrated on an in-house dataset. The rapid and qualitative reconstruction of 4D liver MR has the potential to facilitate online adaptive MR-guided radiotherapy for liver cancer.
Learning Dynamic MRI Reconstruction with Convolutional Network Assisted Reconstruction Swin Transformer
Sep 19, 2023
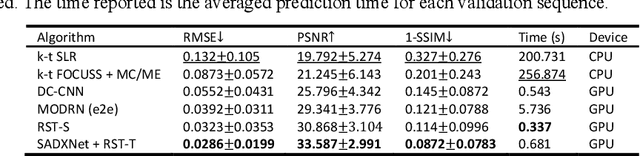

Abstract:Dynamic magnetic resonance imaging (DMRI) is an effective imaging tool for diagnosis tasks that require motion tracking of a certain anatomy. To speed up DMRI acquisition, k-space measurements are commonly undersampled along spatial or spatial-temporal domains. The difficulty of recovering useful information increases with increasing undersampling ratios. Compress sensing was invented for this purpose and has become the most popular method until deep learning (DL) based DMRI reconstruction methods emerged in the past decade. Nevertheless, existing DL networks are still limited in long-range sequential dependency understanding and computational efficiency and are not fully automated. Considering the success of Transformers positional embedding and "swin window" self-attention mechanism in the vision community, especially natural video understanding, we hereby propose a novel architecture named Reconstruction Swin Transformer (RST) for 4D MRI. RST inherits the backbone design of the Video Swin Transformer with a novel reconstruction head introduced to restore pixel-wise intensity. A convolution network called SADXNet is used for rapid initialization of 2D MR frames before RST learning to effectively reduce the model complexity, GPU hardware demand, and training time. Experimental results in the cardiac 4D MR dataset further substantiate the superiority of RST, achieving the lowest RMSE of 0.0286 +/- 0.0199 and 1 - SSIM of 0.0872 +/- 0.0783 on 9 times accelerated validation sequences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge