Accelerated Patient-specific Non-Cartesian MRI Reconstruction using Implicit Neural Representations
Paper and Code
Mar 07, 2025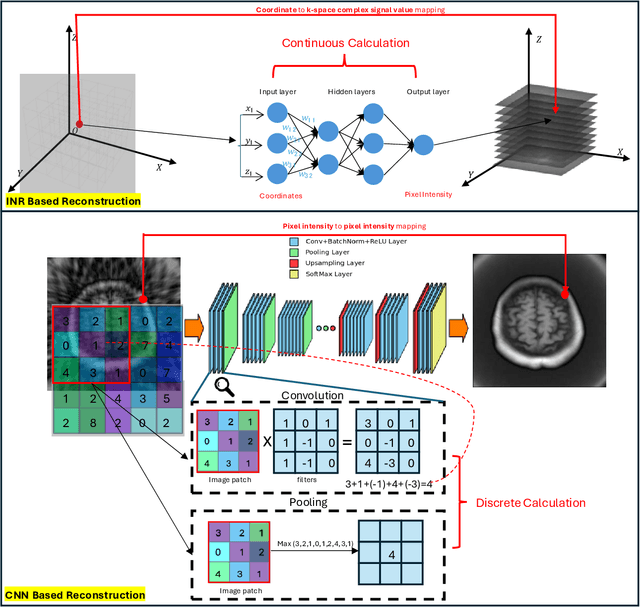
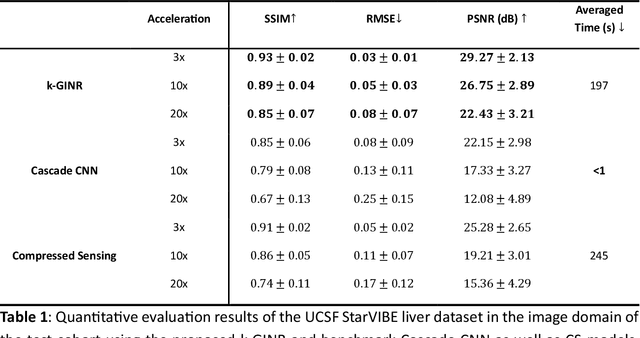
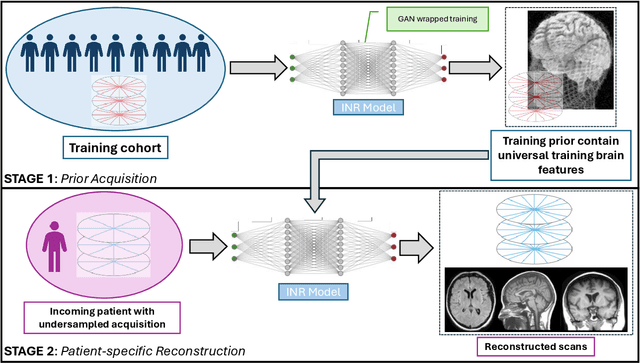
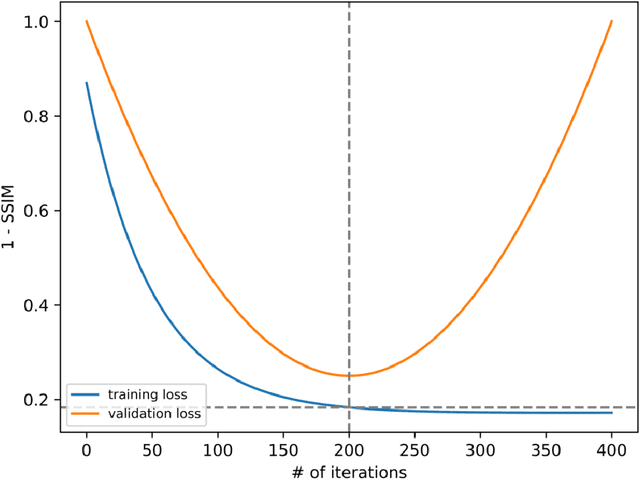
The scanning time for a fully sampled MRI can be undesirably lengthy. Compressed sensing has been developed to minimize image artifacts in accelerated scans, but the required iterative reconstruction is computationally complex and difficult to generalize on new cases. Image-domain-based deep learning methods (e.g., convolutional neural networks) emerged as a faster alternative but face challenges in modeling continuous k-space, a problem amplified with non-Cartesian sampling commonly used in accelerated acquisition. In comparison, implicit neural representations can model continuous signals in the frequency domain and thus are compatible with arbitrary k-space sampling patterns. The current study develops a novel generative-adversarially trained implicit neural representations (k-GINR) for de novo undersampled non-Cartesian k-space reconstruction. k-GINR consists of two stages: 1) supervised training on an existing patient cohort; 2) self-supervised patient-specific optimization. In stage 1, the network is trained with the generative-adversarial network on diverse patients of the same anatomical region supervised by fully sampled acquisition. In stage 2, undersampled k-space data of individual patients is used to tailor the prior-embedded network for patient-specific optimization. The UCSF StarVIBE T1-weighted liver dataset was evaluated on the proposed framework. k-GINR is compared with an image-domain deep learning method, Deep Cascade CNN, and a compressed sensing method. k-GINR consistently outperformed the baselines with a larger performance advantage observed at very high accelerations (e.g., 20 times). k-GINR offers great value for direct non-Cartesian k-space reconstruction for new incoming patients across a wide range of accelerations liver anatomy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge