Debiao Li
Let Distortion Guide Restoration (DGR): A physics-informed learning framework for Prostate Diffusion MRI
Jan 01, 2026Abstract:We present Distortion-Guided Restoration (DGR), a physics-informed hybrid CNN-diffusion framework for acquisition-free correction of severe susceptibility-induced distortions in prostate single-shot EPI diffusion-weighted imaging (DWI). DGR is trained to invert a realistic forward distortion model using large-scale paired distorted and undistorted data synthesized from distortion-free prostate DWI and co-registered T2-weighted images from 410 multi-institutional studies, together with 11 measured B0 field maps from metal-implant cases incorporated into a forward simulator to generate low-b DWI (b = 50 s per mm squared), high-b DWI (b = 1400 s per mm squared), and ADC distortions. The network couples a CNN-based geometric correction module with conditional diffusion refinement under T2-weighted anatomical guidance. On a held-out synthetic validation set (n = 34) using ground-truth simulated distortion fields, DGR achieved higher PSNR and lower NMSE than FSL TOPUP and FUGUE. In 34 real clinical studies with severe distortion, including hip prostheses and marked rectal distension, DGR improved geometric fidelity and increased radiologist-rated image quality and diagnostic confidence. Overall, learning the inverse of a physically simulated forward process provides a practical alternative to acquisition-dependent distortion-correction pipelines for prostate DWI.
EchoPrime: A Multi-Video View-Informed Vision-Language Model for Comprehensive Echocardiography Interpretation
Oct 13, 2024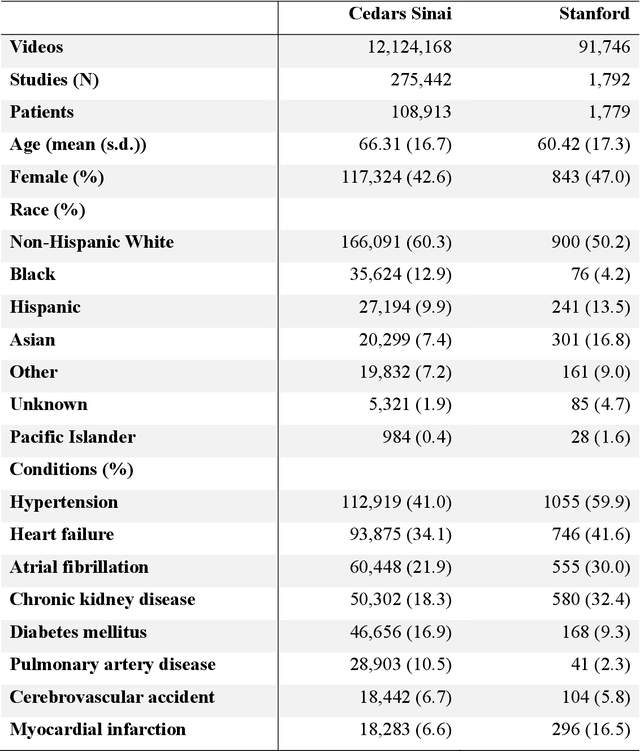
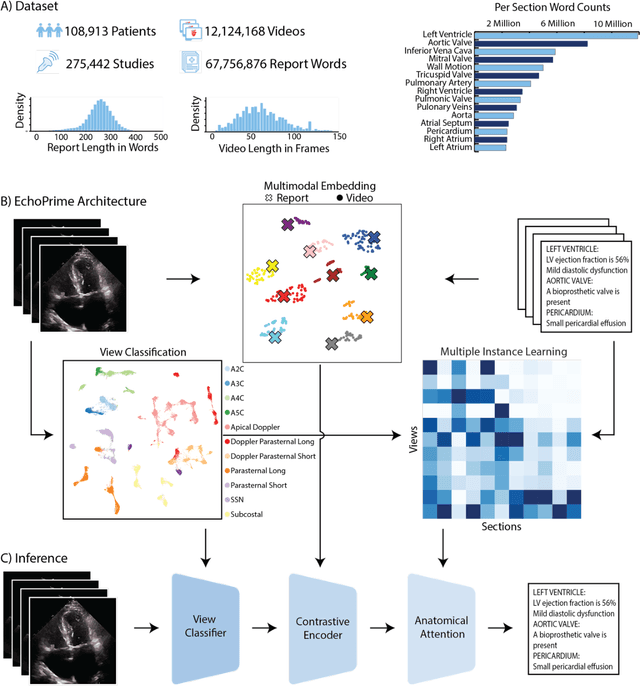
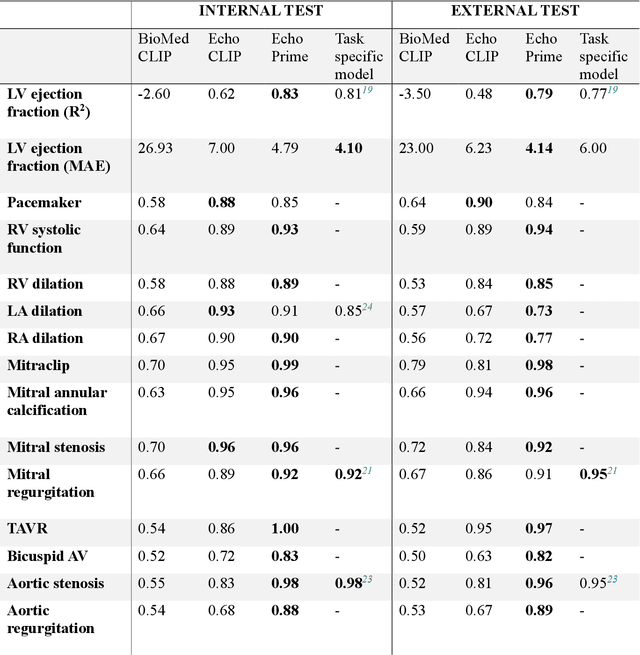
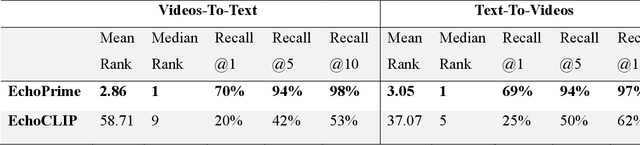
Abstract:Echocardiography is the most widely used cardiac imaging modality, capturing ultrasound video data to assess cardiac structure and function. Artificial intelligence (AI) in echocardiography has the potential to streamline manual tasks and improve reproducibility and precision. However, most echocardiography AI models are single-view, single-task systems that do not synthesize complementary information from multiple views captured during a full exam, and thus lead to limited performance and scope of applications. To address this problem, we introduce EchoPrime, a multi-view, view-informed, video-based vision-language foundation model trained on over 12 million video-report pairs. EchoPrime uses contrastive learning to train a unified embedding model for all standard views in a comprehensive echocardiogram study with representation of both rare and common diseases and diagnoses. EchoPrime then utilizes view-classification and a view-informed anatomic attention model to weight video-specific interpretations that accurately maps the relationship between echocardiographic views and anatomical structures. With retrieval-augmented interpretation, EchoPrime integrates information from all echocardiogram videos in a comprehensive study and performs holistic comprehensive clinical echocardiography interpretation. In datasets from two independent healthcare systems, EchoPrime achieves state-of-the art performance on 23 diverse benchmarks of cardiac form and function, surpassing the performance of both task-specific approaches and prior foundation models. Following rigorous clinical evaluation, EchoPrime can assist physicians in the automated preliminary assessment of comprehensive echocardiography.
Data-Consistent Non-Cartesian Deep Subspace Learning for Efficient Dynamic MR Image Reconstruction
May 03, 2022



Abstract:Non-Cartesian sampling with subspace-constrained image reconstruction is a popular approach to dynamic MRI, but slow iterative reconstruction limits its clinical application. Data-consistent (DC) deep learning can accelerate reconstruction with good image quality, but has not been formulated for non-Cartesian subspace imaging. In this study, we propose a DC non-Cartesian deep subspace learning framework for fast, accurate dynamic MR image reconstruction. Four novel DC formulations are developed and evaluated: two gradient decent approaches, a directly solved approach, and a conjugate gradient approach. We applied a U-Net model with and without DC layers to reconstruct T1-weighted images for cardiac MR Multitasking (an advanced multidimensional imaging method), comparing our results to the iteratively reconstructed reference. Experimental results show that the proposed framework significantly improves reconstruction accuracy over the U-Net model without DC, while significantly accelerating the reconstruction over conventional iterative reconstruction.
Cine Cardiac MRI Motion Artifact Reduction Using a Recurrent Neural Network
Jun 23, 2020



Abstract:Cine cardiac magnetic resonance imaging (MRI) is widely used for diagnosis of cardiac diseases thanks to its ability to present cardiovascular features in excellent contrast. As compared to computed tomography (CT), MRI, however, requires a long scan time, which inevitably induces motion artifacts and causes patients' discomfort. Thus, there has been a strong clinical motivation to develop techniques to reduce both the scan time and motion artifacts. Given its successful applications in other medical imaging tasks such as MRI super-resolution and CT metal artifact reduction, deep learning is a promising approach for cardiac MRI motion artifact reduction. In this paper, we propose a recurrent neural network to simultaneously extract both spatial and temporal features from under-sampled, motion-blurred cine cardiac images for improved image quality. The experimental results demonstrate substantially improved image quality on two clinical test datasets. Also, our method enables data-driven frame interpolation at an enhanced temporal resolution. Compared with existing methods, our deep learning approach gives a superior performance in terms of structural similarity (SSIM) and peak signal-to-noise ratio (PSNR).
MRI Super-Resolution with GAN and 3D Multi-Level DenseNet: Smaller, Faster, and Better
Mar 06, 2020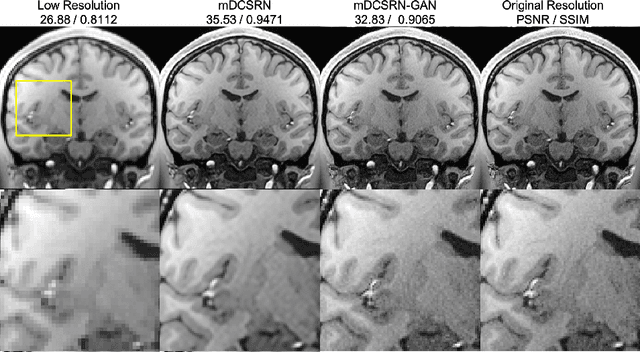
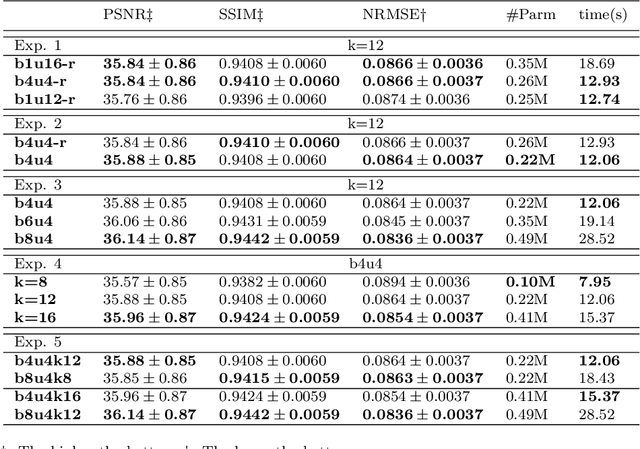
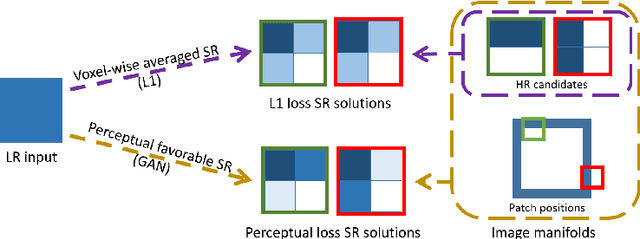
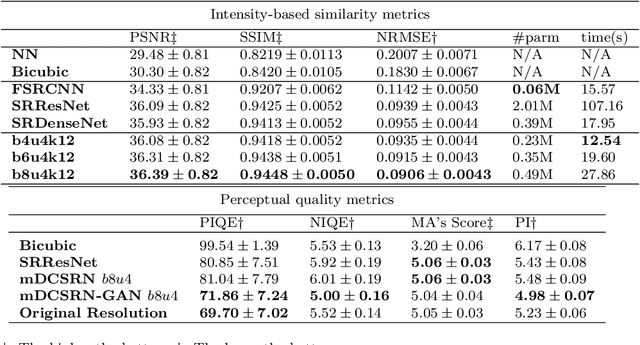
Abstract:High-resolution (HR) magnetic resonance imaging (MRI) provides detailed anatomical information that is critical for diagnosis in the clinical application. However, HR MRI typically comes at the cost of long scan time, small spatial coverage, and low signal-to-noise ratio (SNR). Recent studies showed that with a deep convolutional neural network (CNN), HR generic images could be recovered from low-resolution (LR) inputs via single image super-resolution (SISR) approaches. Additionally, previous works have shown that a deep 3D CNN can generate high-quality SR MRIs by using learned image priors. However, 3D CNN with deep structures, have a large number of parameters and are computationally expensive. In this paper, we propose a novel 3D CNN architecture, namely a multi-level densely connected super-resolution network (mDCSRN), which is light-weight, fast and accurate. We also show that with the generative adversarial network (GAN)-guided training, the mDCSRN-GAN provides appealing sharp SR images with rich texture details that are highly comparable with the referenced HR images. Our results from experiments on a large public dataset with 1,113 subjects showed that this new architecture outperformed other popular deep learning methods in recovering 4x resolution-downgraded images in both quality and speed.
Fully Automated Multi-Organ Segmentation in Abdominal Magnetic Resonance Imaging with Deep Neural Networks
Dec 23, 2019



Abstract:Segmentation of multiple organs-at-risk (OARs) is essential for radiation therapy treatment planning and other clinical applications. We developed an Automated deep Learning-based Abdominal Multi-Organ segmentation (ALAMO) framework based on 2D U-net and a densely connected network structure with tailored design in data augmentation and training procedures such as deep connection, auxiliary supervision, and multi-view. The model takes in multi-slice MR images and generates the output of segmentation results. Three-Tesla T1 VIBE (Volumetric Interpolated Breath-hold Examination) images of 102 subjects were collected and used in our study. Ten OARs were studied, including the liver, spleen, pancreas, left/right kidneys, stomach, duodenum, small intestine, spinal cord, and vertebral bodies. Two radiologists manually labeled and obtained the consensus contours as the ground-truth. In the complete cohort of 102, 20 samples were held out for independent testing, and the rest were used for training and validation. The performance was measured using volume overlapping and surface distance. The ALAMO framework generated segmentation labels in good agreement with the manual results. Specifically, among the 10 OARs, 9 achieved high Dice Similarity Coefficients (DSCs) in the range of 0.87-0.96, except for the duodenum with a DSC of 0.80. The inference completes within one minute for a 3D volume of 320x288x180. Overall, the ALAMO model matches the state-of-the-art performance. The proposed ALAMO framework allows for fully automated abdominal MR segmentation with high accuracy and low memory and computation time demands.
Deep learning within a priori temporal feature spaces for large-scale dynamic MR image reconstruction: Application to 5-D cardiac MR Multitasking
Oct 02, 2019



Abstract:High spatiotemporal resolution dynamic magnetic resonance imaging (MRI) is a powerful clinical tool for imaging moving structures as well as to reveal and quantify other physical and physiological dynamics. The low speed of MRI necessitates acceleration methods such as deep learning reconstruction from under-sampled data. However, the massive size of many dynamic MRI problems prevents deep learning networks from directly exploiting global temporal relationships. In this work, we show that by applying deep neural networks inside a priori calculated temporal feature spaces, we enable deep learning reconstruction with global temporal modeling even for image sequences with >40,000 frames. One proposed variation of our approach using dilated multi-level Densely Connected Network (mDCN) speeds up feature space coordinate calculation by 3000x compared to conventional iterative methods, from 20 minutes to 0.39 seconds. Thus, the combination of low-rank tensor and deep learning models not only makes large-scale dynamic MRI feasible but also practical for routine clinical application.
Efficient and Accurate MRI Super-Resolution using a Generative Adversarial Network and 3D Multi-Level Densely Connected Network
Jun 09, 2018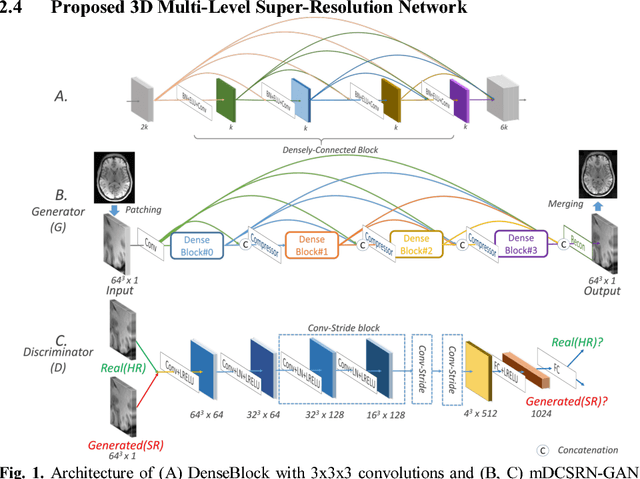
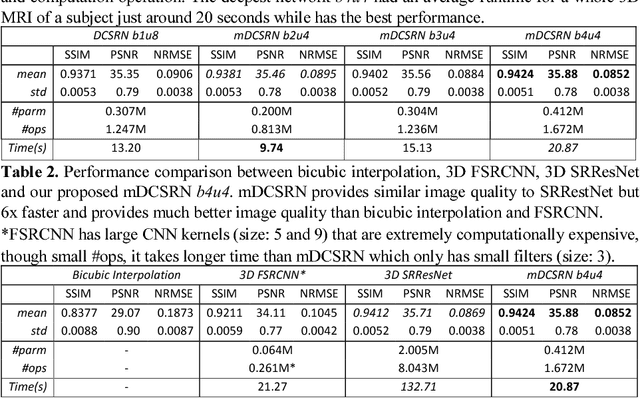
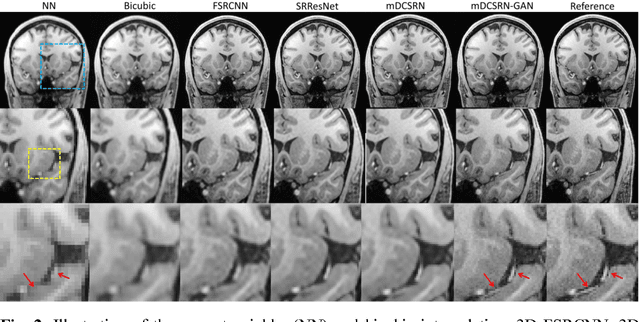
Abstract:High-resolution (HR) magnetic resonance images (MRI) provide detailed anatomical information important for clinical application and quantitative image analysis. However, HR MRI conventionally comes at the cost of longer scan time, smaller spatial coverage, and lower signal-to-noise ratio (SNR). Recent studies have shown that single image super-resolution (SISR), a technique to recover HR details from one single low-resolution (LR) input image, could provide high-quality image details with the help of advanced deep convolutional neural networks (CNN). However, deep neural networks consume memory heavily and run slowly, especially in 3D settings. In this paper, we propose a novel 3D neural network design, namely a multi-level densely connected super-resolution network (mDCSRN) with generative adversarial network (GAN)-guided training. The mDCSRN quickly trains and inferences and the GAN promotes realistic output hardly distinguishable from original HR images. Our results from experiments on a dataset with 1,113 subjects show that our new architecture beats other popular deep learning methods in recovering 4x resolution-downgraded im-ages and runs 6x faster.
Calcium Removal From Cardiac CT Images Using Deep Convolutional Neural Network
Feb 20, 2018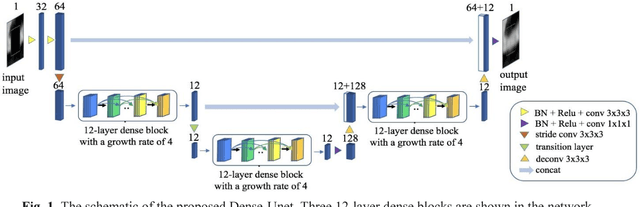
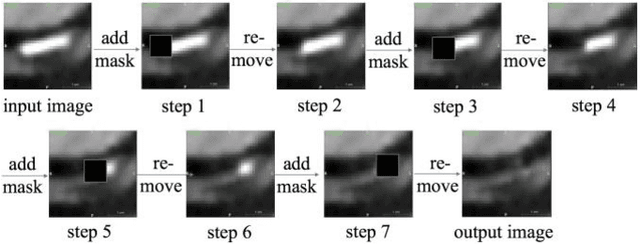
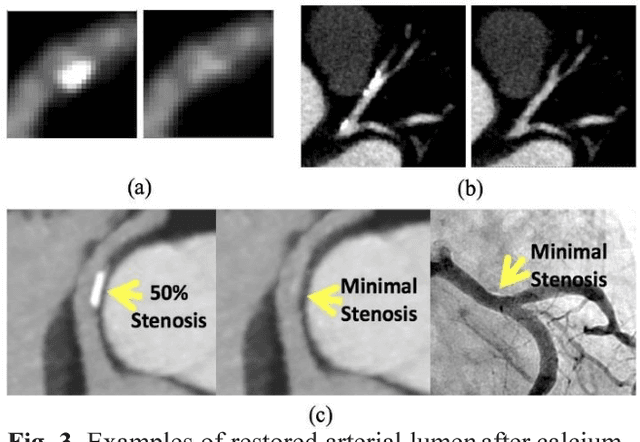

Abstract:Coronary calcium causes beam hardening and blooming artifacts on cardiac computed tomography angiography (CTA) images, which lead to overestimation of lumen stenosis and reduction of diagnostic specificity. To properly remove coronary calcification and restore arterial lumen precisely, we propose a machine learning-based method with a multi-step inpainting process. We developed a new network configuration, Dense-Unet, to achieve optimal performance with low computational cost. Results after the calcium removal process were validated by comparing with gold-standard X-ray angiography. Our results demonstrated that removing coronary calcification from images with the proposed approach was feasible, and may potentially improve the diagnostic accuracy of CTA.
Brain MRI Super Resolution Using 3D Deep Densely Connected Neural Networks
Jan 08, 2018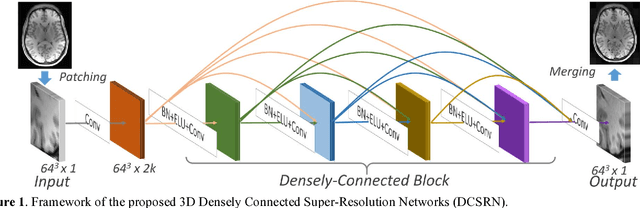
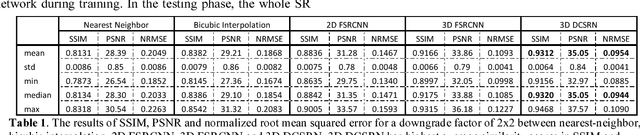
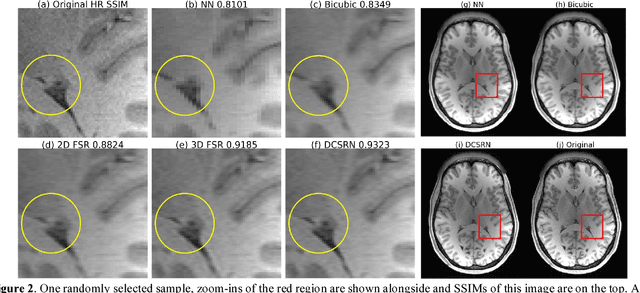
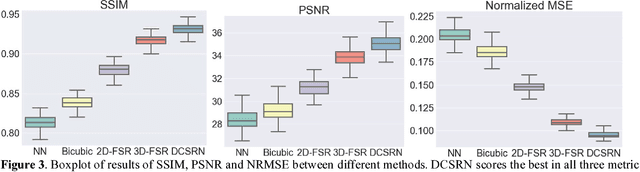
Abstract:Magnetic resonance image (MRI) in high spatial resolution provides detailed anatomical information and is often necessary for accurate quantitative analysis. However, high spatial resolution typically comes at the expense of longer scan time, less spatial coverage, and lower signal to noise ratio (SNR). Single Image Super-Resolution (SISR), a technique aimed to restore high-resolution (HR) details from one single low-resolution (LR) input image, has been improved dramatically by recent breakthroughs in deep learning. In this paper, we introduce a new neural network architecture, 3D Densely Connected Super-Resolution Networks (DCSRN) to restore HR features of structural brain MR images. Through experiments on a dataset with 1,113 subjects, we demonstrate that our network outperforms bicubic interpolation as well as other deep learning methods in restoring 4x resolution-reduced images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge