Bryan He
Stanford Sleep Bench: Evaluating Polysomnography Pre-training Methods for Sleep Foundation Models
Dec 10, 2025Abstract:Polysomnography (PSG), the gold standard test for sleep analysis, generates vast amounts of multimodal clinical data, presenting an opportunity to leverage self-supervised representation learning (SSRL) for pre-training foundation models to enhance sleep analysis. However, progress in sleep foundation models is hindered by two key limitations: (1) the lack of a shared dataset and benchmark with diverse tasks for training and evaluation, and (2) the absence of a systematic evaluation of SSRL approaches across sleep-related tasks. To address these gaps, we introduce Stanford Sleep Bench, a large-scale PSG dataset comprising 17,467 recordings totaling over 163,000 hours from a major sleep clinic, including 13 clinical disease prediction tasks alongside canonical sleep-related tasks such as sleep staging, apnea diagnosis, and age estimation. We systematically evaluate SSRL pre-training methods on Stanford Sleep Bench, assessing downstream performance across four tasks: sleep staging, apnea diagnosis, age estimation, and disease and mortality prediction. Our results show that multiple pretraining methods achieve comparable performance for sleep staging, apnea diagnosis, and age estimation. However, for mortality and disease prediction, contrastive learning significantly outperforms other approaches while also converging faster during pretraining. To facilitate reproducibility and advance sleep research, we will release Stanford Sleep Bench along with pretrained model weights, training pipelines, and evaluation code.
Medical Image De-Identification Benchmark Challenge
Jul 31, 2025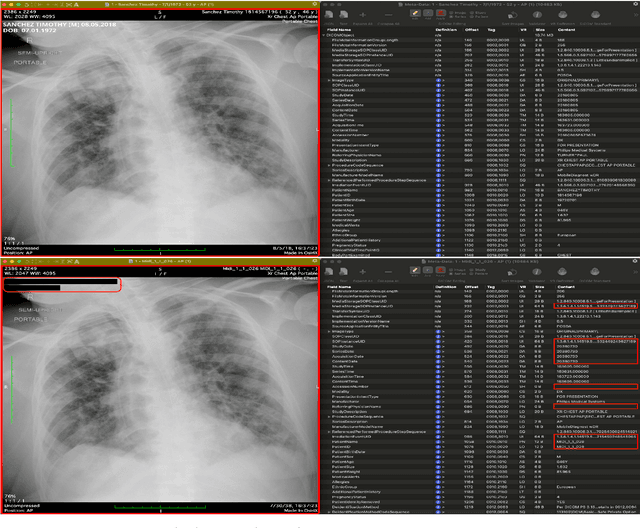
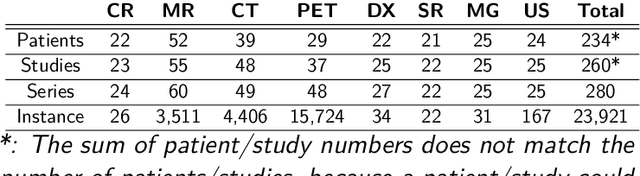
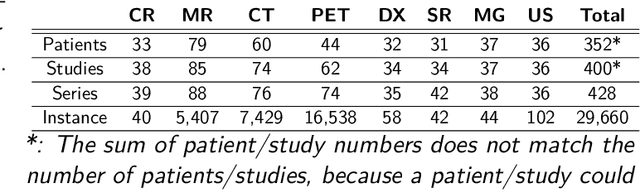
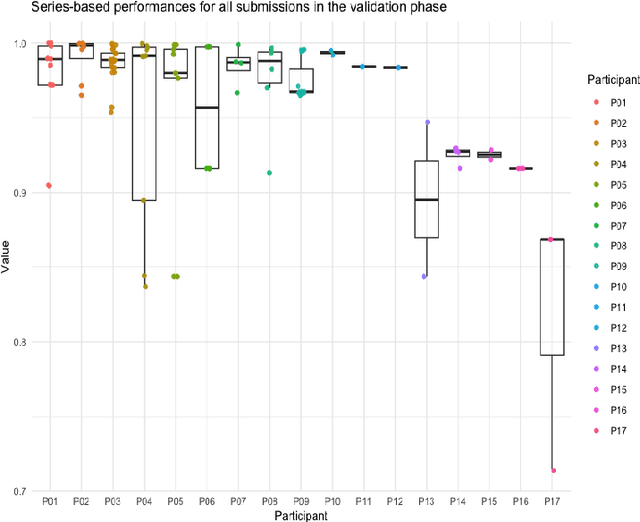
Abstract:The de-identification (deID) of protected health information (PHI) and personally identifiable information (PII) is a fundamental requirement for sharing medical images, particularly through public repositories, to ensure compliance with patient privacy laws. In addition, preservation of non-PHI metadata to inform and enable downstream development of imaging artificial intelligence (AI) is an important consideration in biomedical research. The goal of MIDI-B was to provide a standardized platform for benchmarking of DICOM image deID tools based on a set of rules conformant to the HIPAA Safe Harbor regulation, the DICOM Attribute Confidentiality Profiles, and best practices in preservation of research-critical metadata, as defined by The Cancer Imaging Archive (TCIA). The challenge employed a large, diverse, multi-center, and multi-modality set of real de-identified radiology images with synthetic PHI/PII inserted. The MIDI-B Challenge consisted of three phases: training, validation, and test. Eighty individuals registered for the challenge. In the training phase, we encouraged participants to tune their algorithms using their in-house or public data. The validation and test phases utilized the DICOM images containing synthetic identifiers (of 216 and 322 subjects, respectively). Ten teams successfully completed the test phase of the challenge. To measure success of a rule-based approach to image deID, scores were computed as the percentage of correct actions from the total number of required actions. The scores ranged from 97.91% to 99.93%. Participants employed a variety of open-source and proprietary tools with customized configurations, large language models, and optical character recognition (OCR). In this paper we provide a comprehensive report on the MIDI-B Challenge's design, implementation, results, and lessons learned.
How Well Can General Vision-Language Models Learn Medicine By Watching Public Educational Videos?
Apr 19, 2025Abstract:Publicly available biomedical videos, such as those on YouTube, serve as valuable educational resources for medical students. Unlike standard machine learning datasets, these videos are designed for human learners, often mixing medical imagery with narration, explanatory diagrams, and contextual framing. In this work, we investigate whether such pedagogically rich, yet non-standardized and heterogeneous videos can effectively teach general-domain vision-language models biomedical knowledge. To this end, we introduce OpenBiomedVi, a biomedical video instruction tuning dataset comprising 1031 hours of video-caption and Q/A pairs, curated through a multi-step human-in-the-loop pipeline. Diverse biomedical video datasets are rare, and OpenBiomedVid fills an important gap by providing instruction-style supervision grounded in real-world educational content. Surprisingly, despite the informal and heterogeneous nature of these videos, the fine-tuned Qwen-2-VL models exhibit substantial performance improvements across most benchmarks. The 2B model achieves gains of 98.7% on video tasks, 71.2% on image tasks, and 0.2% on text tasks. The 7B model shows improvements of 37.09% on video and 11.2% on image tasks, with a slight degradation of 2.7% on text tasks compared to their respective base models. To address the lack of standardized biomedical video evaluation datasets, we also introduce two new expert curated benchmarks, MIMICEchoQA and SurgeryVideoQA. On these benchmarks, the 2B model achieves gains of 99.1% and 98.1%, while the 7B model shows gains of 22.5% and 52.1%, respectively, demonstrating the models' ability to generalize and perform biomedical video understanding on cleaner and more standardized datasets than those seen during training. These results suggest that educational videos created for human learning offer a surprisingly effective training signal for biomedical VLMs.
EchoPrime: A Multi-Video View-Informed Vision-Language Model for Comprehensive Echocardiography Interpretation
Oct 13, 2024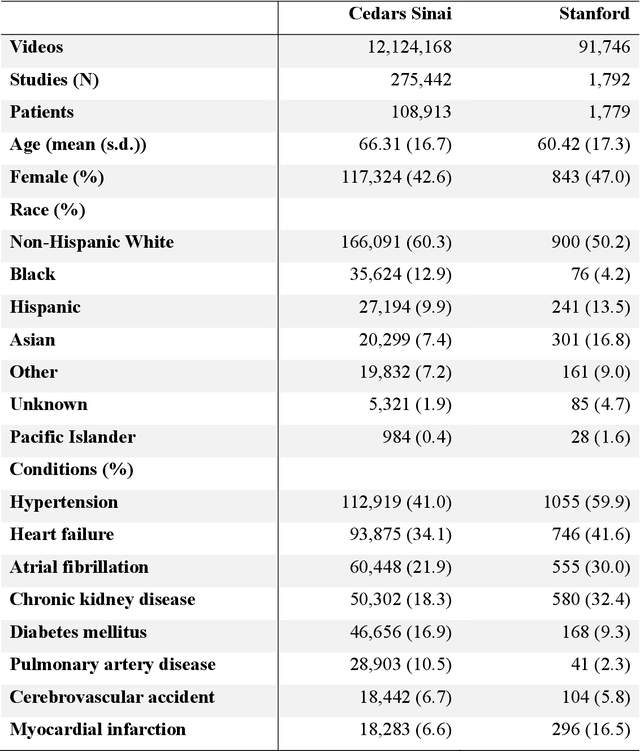
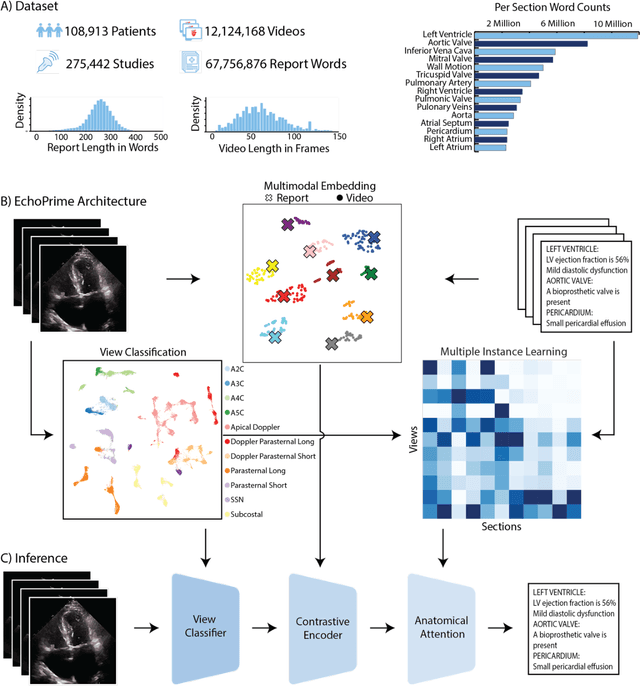
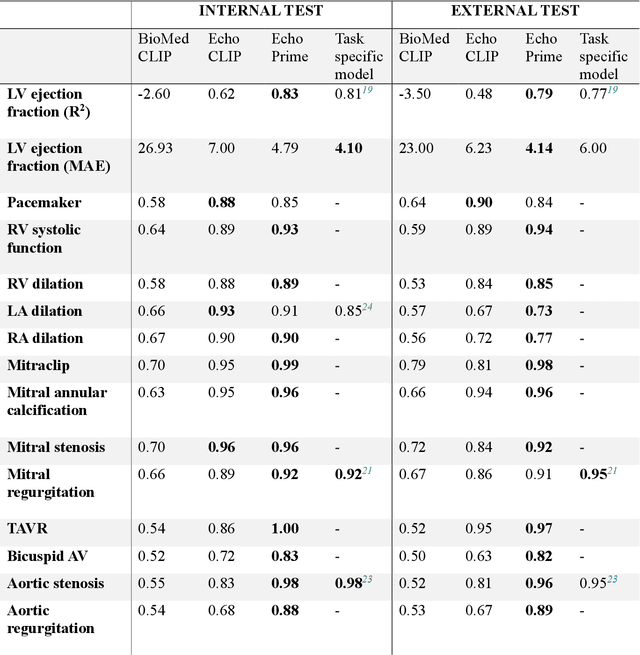
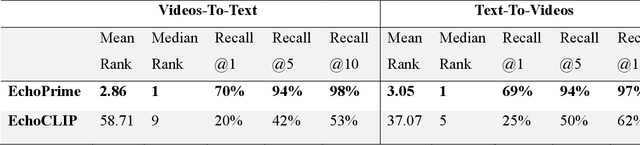
Abstract:Echocardiography is the most widely used cardiac imaging modality, capturing ultrasound video data to assess cardiac structure and function. Artificial intelligence (AI) in echocardiography has the potential to streamline manual tasks and improve reproducibility and precision. However, most echocardiography AI models are single-view, single-task systems that do not synthesize complementary information from multiple views captured during a full exam, and thus lead to limited performance and scope of applications. To address this problem, we introduce EchoPrime, a multi-view, view-informed, video-based vision-language foundation model trained on over 12 million video-report pairs. EchoPrime uses contrastive learning to train a unified embedding model for all standard views in a comprehensive echocardiogram study with representation of both rare and common diseases and diagnoses. EchoPrime then utilizes view-classification and a view-informed anatomic attention model to weight video-specific interpretations that accurately maps the relationship between echocardiographic views and anatomical structures. With retrieval-augmented interpretation, EchoPrime integrates information from all echocardiogram videos in a comprehensive study and performs holistic comprehensive clinical echocardiography interpretation. In datasets from two independent healthcare systems, EchoPrime achieves state-of-the art performance on 23 diverse benchmarks of cardiac form and function, surpassing the performance of both task-specific approaches and prior foundation models. Following rigorous clinical evaluation, EchoPrime can assist physicians in the automated preliminary assessment of comprehensive echocardiography.
SleepFM: Multi-modal Representation Learning for Sleep Across Brain Activity, ECG and Respiratory Signals
May 28, 2024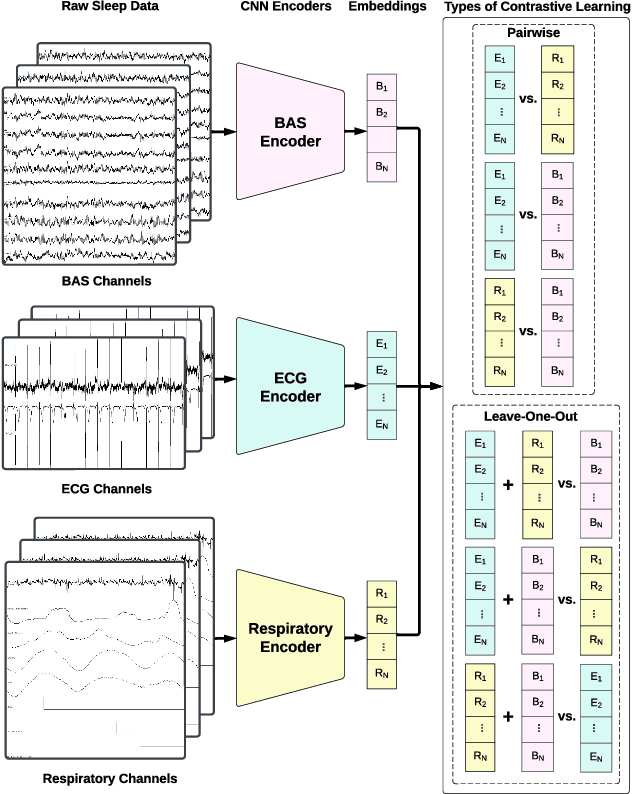
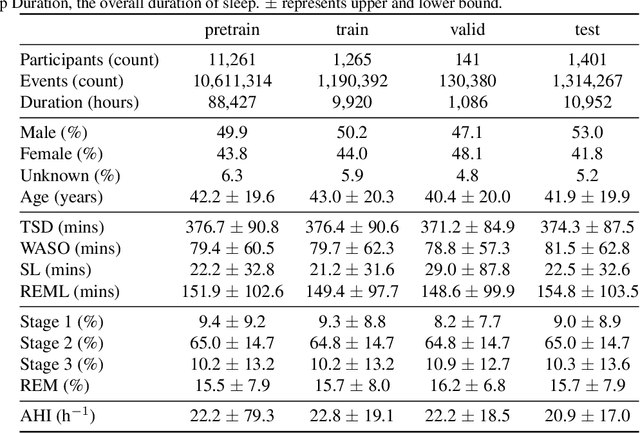
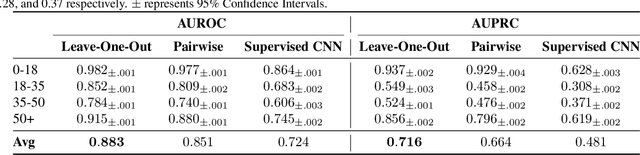
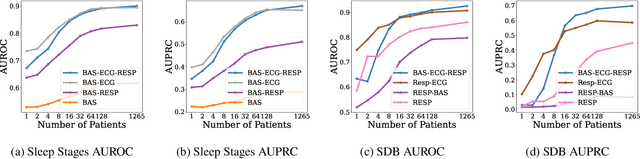
Abstract:Sleep is a complex physiological process evaluated through various modalities recording electrical brain, cardiac, and respiratory activities. We curate a large polysomnography dataset from over 14,000 participants comprising over 100,000 hours of multi-modal sleep recordings. Leveraging this extensive dataset, we developed SleepFM, the first multi-modal foundation model for sleep analysis. We show that a novel leave-one-out approach for contrastive learning significantly improves downstream task performance compared to representations from standard pairwise contrastive learning. A logistic regression model trained on SleepFM's learned embeddings outperforms an end-to-end trained convolutional neural network (CNN) on sleep stage classification (macro AUROC 0.88 vs 0.72 and macro AUPRC 0.72 vs 0.48) and sleep disordered breathing detection (AUROC 0.85 vs 0.69 and AUPRC 0.77 vs 0.61). Notably, the learned embeddings achieve 48% top-1 average accuracy in retrieving the corresponding recording clips of other modalities from 90,000 candidates. This work demonstrates the value of holistic multi-modal sleep modeling to fully capture the richness of sleep recordings. SleepFM is open source and available at https://github.com/rthapa84/sleepfm-codebase.
Deep Learning Discovery of Demographic Biomarkers in Echocardiography
Jul 13, 2022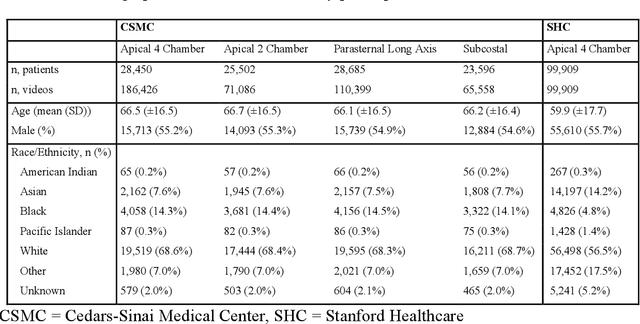
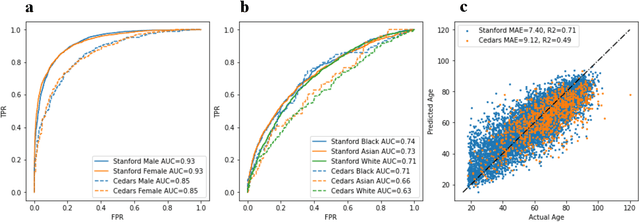
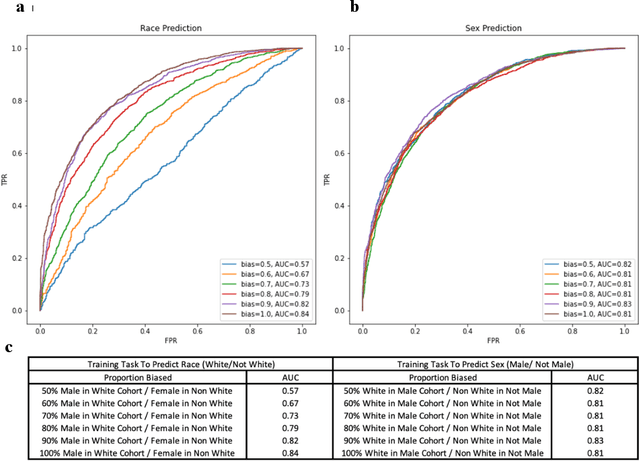
Abstract:Deep learning has been shown to accurately assess 'hidden' phenotypes and predict biomarkers from medical imaging beyond traditional clinician interpretation of medical imaging. Given the black box nature of artificial intelligence (AI) models, caution should be exercised in applying models to healthcare as prediction tasks might be short-cut by differences in demographics across disease and patient populations. Using large echocardiography datasets from two healthcare systems, we test whether it is possible to predict age, race, and sex from cardiac ultrasound images using deep learning algorithms and assess the impact of varying confounding variables. We trained video-based convolutional neural networks to predict age, sex, and race. We found that deep learning models were able to identify age and sex, while unable to reliably predict race. Without considering confounding differences between categories, the AI model predicted sex with an AUC of 0.85 (95% CI 0.84 - 0.86), age with a mean absolute error of 9.12 years (95% CI 9.00 - 9.25), and race with AUCs ranging from 0.63 - 0.71. When predicting race, we show that tuning the proportion of a confounding variable (sex) in the training data significantly impacts model AUC (ranging from 0.57 to 0.84), while in training a sex prediction model, tuning a confounder (race) did not substantially change AUC (0.81 - 0.83). This suggests a significant proportion of the model's performance on predicting race could come from confounding features being detected by AI. Further work remains to identify the particular imaging features that associate with demographic information and to better understand the risks of demographic identification in medical AI as it pertains to potentially perpetuating bias and disparities.
CloudPred: Predicting Patient Phenotypes From Single-cell RNA-seq
Oct 13, 2021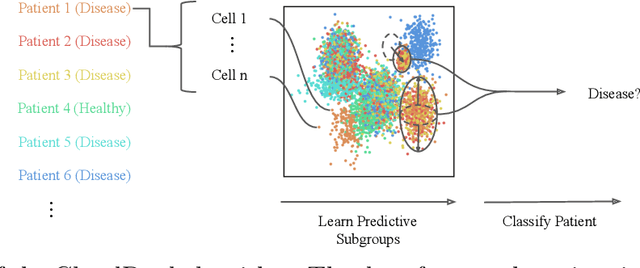
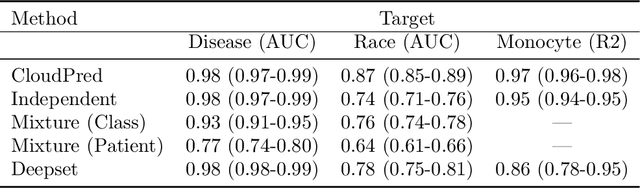
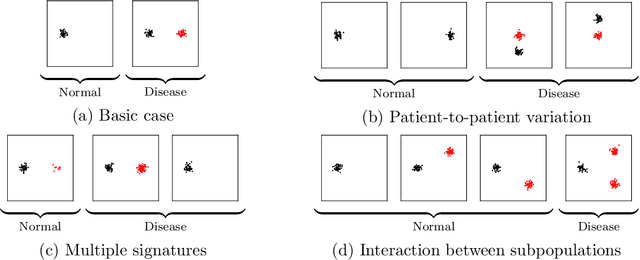
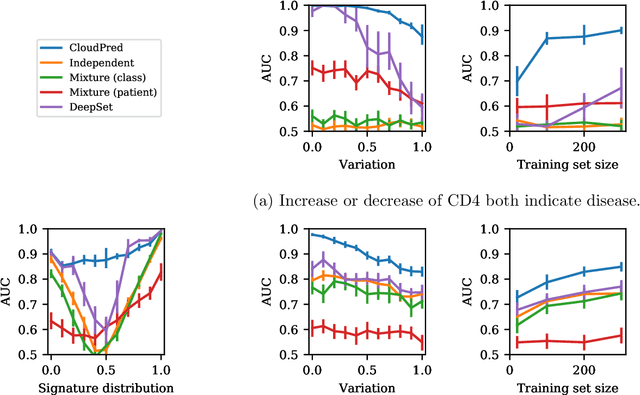
Abstract:Single-cell RNA sequencing (scRNA-seq) has the potential to provide powerful, high-resolution signatures to inform disease prognosis and precision medicine. This paper takes an important first step towards this goal by developing an interpretable machine learning algorithm, CloudPred, to predict individuals' disease phenotypes from their scRNA-seq data. Predicting phenotype from scRNA-seq is challenging for standard machine learning methods -- the number of cells measured can vary by orders of magnitude across individuals and the cell populations are also highly heterogeneous. Typical analysis creates pseudo-bulk samples which are biased toward prior annotations and also lose the single cell resolution. CloudPred addresses these challenges via a novel end-to-end differentiable learning algorithm which is coupled with a biologically informed mixture of cell types model. CloudPred automatically infers the cell subpopulation that are salient for the phenotype without prior annotations. We developed a systematic simulation platform to evaluate the performance of CloudPred and several alternative methods we propose, and find that CloudPred outperforms the alternative methods across several settings. We further validated CloudPred on a real scRNA-seq dataset of 142 lupus patients and controls. CloudPred achieves AUROC of 0.98 while identifying a specific subpopulation of CD4 T cells whose presence is highly indicative of lupus. CloudPred is a powerful new framework to predict clinical phenotypes from scRNA-seq data and to identify relevant cells.
High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy with Cardiovascular Deep Learning
Jun 23, 2021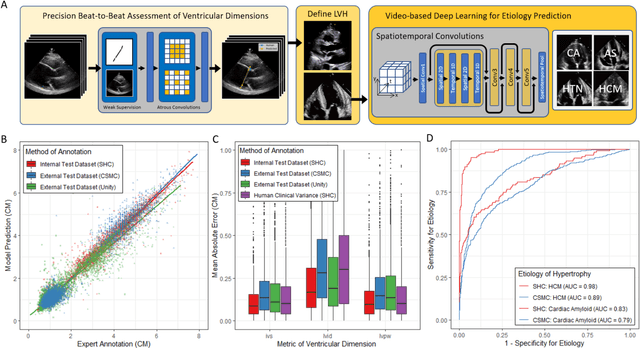
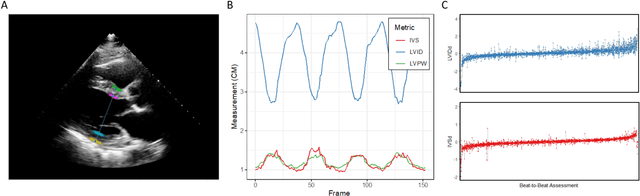
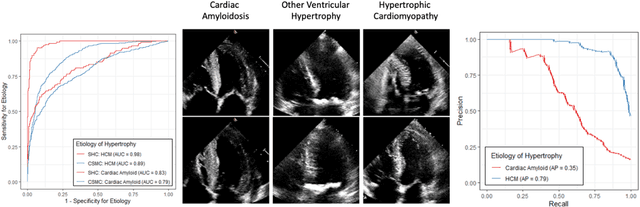
Abstract:Left ventricular hypertrophy (LVH) results from chronic remodeling caused by a broad range of systemic and cardiovascular disease including hypertension, aortic stenosis, hypertrophic cardiomyopathy, and cardiac amyloidosis. Early detection and characterization of LVH can significantly impact patient care but is limited by under-recognition of hypertrophy, measurement error and variability, and difficulty differentiating etiologies of LVH. To overcome this challenge, we present EchoNet-LVH - a deep learning workflow that automatically quantifies ventricular hypertrophy with precision equal to human experts and predicts etiology of LVH. Trained on 28,201 echocardiogram videos, our model accurately measures intraventricular wall thickness (mean absolute error [MAE] 1.4mm, 95% CI 1.2-1.5mm), left ventricular diameter (MAE 2.4mm, 95% CI 2.2-2.6mm), and posterior wall thickness (MAE 1.2mm, 95% CI 1.1-1.3mm) and classifies cardiac amyloidosis (area under the curve of 0.83) and hypertrophic cardiomyopathy (AUC 0.98) from other etiologies of LVH. In external datasets from independent domestic and international healthcare systems, EchoNet-LVH accurately quantified ventricular parameters (R2 of 0.96 and 0.90 respectively) and detected cardiac amyloidosis (AUC 0.79) and hypertrophic cardiomyopathy (AUC 0.89) on the domestic external validation site. Leveraging measurements across multiple heart beats, our model can more accurately identify subtle changes in LV geometry and its causal etiologies. Compared to human experts, EchoNet-LVH is fully automated, allowing for reproducible, precise measurements, and lays the foundation for precision diagnosis of cardiac hypertrophy. As a resource to promote further innovation, we also make publicly available a large dataset of 23,212 annotated echocardiogram videos.
Diversity Breeds Innovation With Discounted Impact and Recognition
Sep 04, 2019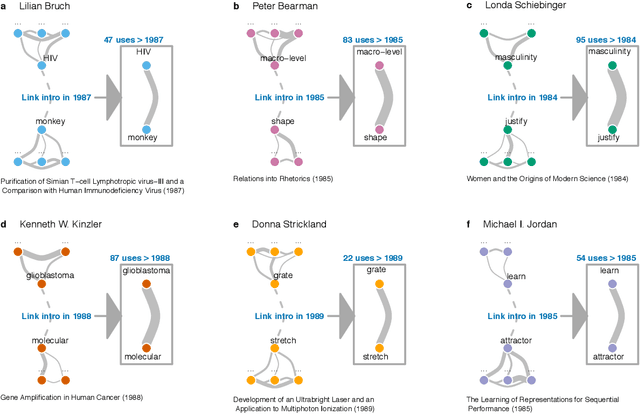
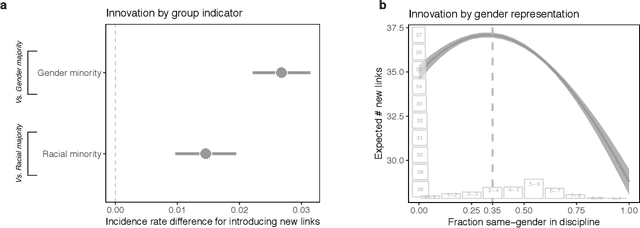

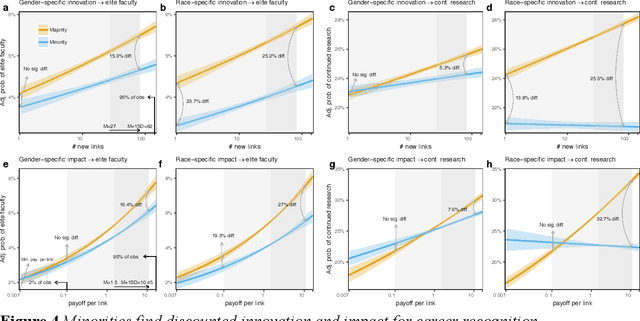
Abstract:Prior work poses a diversity paradox for science. Diversity breeds scientific innovation, and yet, diverse individuals have less successful scientific careers. But if diversity is good for innovation, why is science not rewarding diversity? We answer this question by utilizing a near-population of ~1.03 million US doctoral recipients from 1980-2015 and their careers into publishing and faculty roles. The article uses text analysis and machine learning techniques to answer a series of questions: How can we detect scientific innovation? Does diversity breed innovation? And are the innovations of diverse individuals adopted and rewarded? Our analyses show that underrepresented groups produce higher rates of scientific novelty. However, their novel contributions are discounted: e.g., innovations by gender minorities are taken up by other scholars at lower rates than innovations by gender majorities, and innovations by gender and racial minorities result in fewer academic positions. This suggests an unfair system in which diverse individuals innovate, but their innovations are disproportionately ignored and fail to convert into career success at the same rate as majority groups. In sum, there may be an unwarranted reproduction of stratification in academic careers that discounts diversity's role in innovation and partly explains the underrepresentation of some groups in academia.
Minimizing Close-k Aggregate Loss Improves Classification
Nov 03, 2018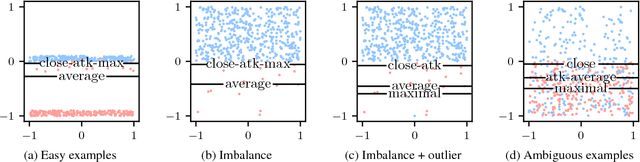
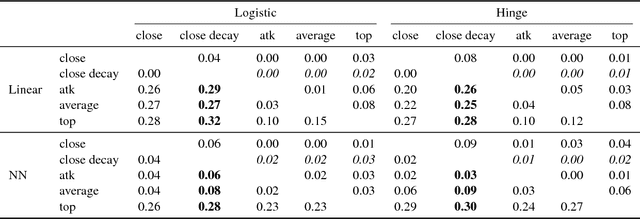
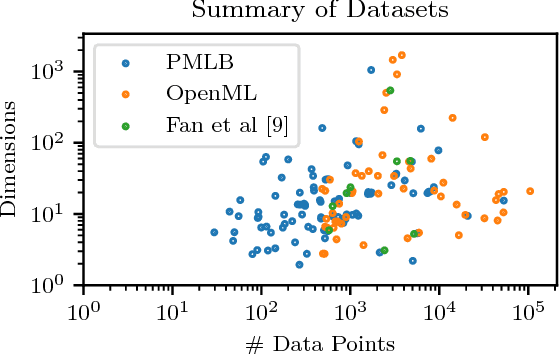

Abstract:In classification, the de facto method for aggregating individual losses is the average loss. When the actual metric of interest is 0-1 loss, it is common to minimize the average surrogate loss for some well-behaved (e.g. convex) surrogate. Recently, several other aggregate losses such as the maximal loss and average top-$k$ loss were proposed as alternative objectives to address shortcomings of the average loss. However, we identify common classification settings, e.g. the data is imbalanced, has too many easy or ambiguous examples, etc., when average, maximal and average top-$k$ all suffer from suboptimal decision boundaries, even on an infinitely large training set. To address this problem, we propose a new classification objective called the close-$k$ aggregate loss, where we adaptively minimize the loss for points close to the decision boundary. We provide theoretical guarantees for the 0-1 accuracy when we optimize close-$k$ aggregate loss. We also conduct systematic experiments across the PMLB and OpenML benchmark datasets. Close-$k$ achieves significant gains in 0-1 test accuracy, improvements of $\geq 2$% and $p<0.05$, in over 25% of the datasets compared to average, maximal and average top-$k$. In contrast, the previous aggregate losses outperformed close-$k$ in less than 2% of the datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge