Angela Zhang
MedCaseReasoning: Evaluating and learning diagnostic reasoning from clinical case reports
May 16, 2025Abstract:Doctors and patients alike increasingly use Large Language Models (LLMs) to diagnose clinical cases. However, unlike domains such as math or coding, where correctness can be objectively defined by the final answer, medical diagnosis requires both the outcome and the reasoning process to be accurate. Currently, widely used medical benchmarks like MedQA and MMLU assess only accuracy in the final answer, overlooking the quality and faithfulness of the clinical reasoning process. To address this limitation, we introduce MedCaseReasoning, the first open-access dataset for evaluating LLMs on their ability to align with clinician-authored diagnostic reasoning. The dataset includes 14,489 diagnostic question-and-answer cases, each paired with detailed reasoning statements derived from open-access medical case reports. We evaluate state-of-the-art reasoning LLMs on MedCaseReasoning and find significant shortcomings in their diagnoses and reasoning: for instance, the top-performing open-source model, DeepSeek-R1, achieves only 48% 10-shot diagnostic accuracy and mentions only 64% of the clinician reasoning statements (recall). However, we demonstrate that fine-tuning LLMs on the reasoning traces derived from MedCaseReasoning significantly improves diagnostic accuracy and clinical reasoning recall by an average relative gain of 29% and 41%, respectively. The open-source dataset, code, and models are available at https://github.com/kevinwu23/Stanford-MedCaseReasoning.
Disentangling Reasoning and Knowledge in Medical Large Language Models
May 16, 2025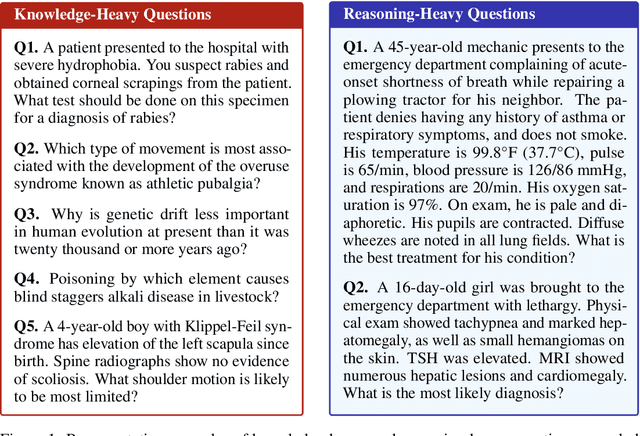
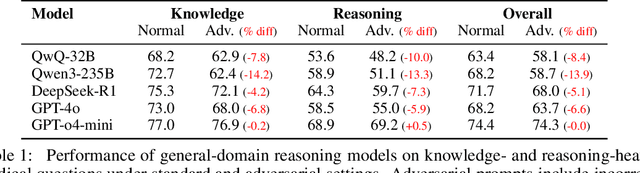
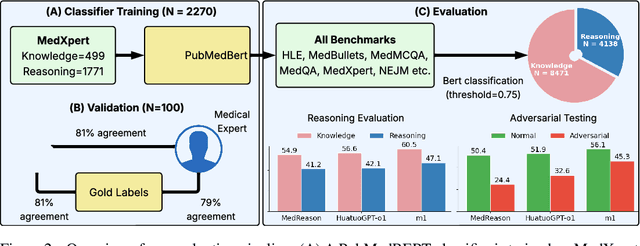
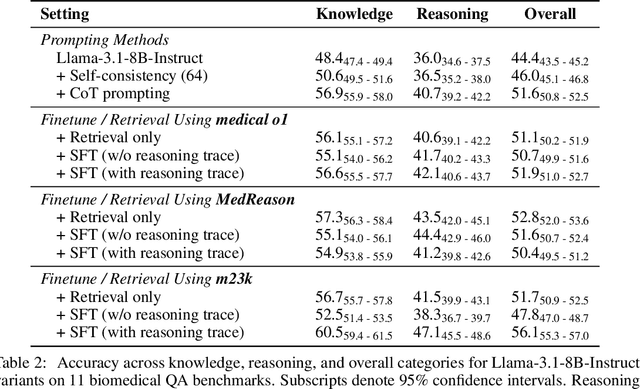
Abstract:Medical reasoning in large language models (LLMs) aims to emulate clinicians' diagnostic thinking, but current benchmarks such as MedQA-USMLE, MedMCQA, and PubMedQA often mix reasoning with factual recall. We address this by separating 11 biomedical QA benchmarks into reasoning- and knowledge-focused subsets using a PubMedBERT classifier that reaches 81 percent accuracy, comparable to human performance. Our analysis shows that only 32.8 percent of questions require complex reasoning. We evaluate biomedical models (HuatuoGPT-o1, MedReason, m1) and general-domain models (DeepSeek-R1, o4-mini, Qwen3), finding consistent gaps between knowledge and reasoning performance. For example, m1 scores 60.5 on knowledge but only 47.1 on reasoning. In adversarial tests where models are misled with incorrect initial reasoning, biomedical models degrade sharply, while larger or RL-trained general models show more robustness. To address this, we train BioMed-R1 using fine-tuning and reinforcement learning on reasoning-heavy examples. It achieves the strongest performance among similarly sized models. Further gains may come from incorporating clinical case reports and training with adversarial and backtracking scenarios.
How Well Can General Vision-Language Models Learn Medicine By Watching Public Educational Videos?
Apr 19, 2025Abstract:Publicly available biomedical videos, such as those on YouTube, serve as valuable educational resources for medical students. Unlike standard machine learning datasets, these videos are designed for human learners, often mixing medical imagery with narration, explanatory diagrams, and contextual framing. In this work, we investigate whether such pedagogically rich, yet non-standardized and heterogeneous videos can effectively teach general-domain vision-language models biomedical knowledge. To this end, we introduce OpenBiomedVi, a biomedical video instruction tuning dataset comprising 1031 hours of video-caption and Q/A pairs, curated through a multi-step human-in-the-loop pipeline. Diverse biomedical video datasets are rare, and OpenBiomedVid fills an important gap by providing instruction-style supervision grounded in real-world educational content. Surprisingly, despite the informal and heterogeneous nature of these videos, the fine-tuned Qwen-2-VL models exhibit substantial performance improvements across most benchmarks. The 2B model achieves gains of 98.7% on video tasks, 71.2% on image tasks, and 0.2% on text tasks. The 7B model shows improvements of 37.09% on video and 11.2% on image tasks, with a slight degradation of 2.7% on text tasks compared to their respective base models. To address the lack of standardized biomedical video evaluation datasets, we also introduce two new expert curated benchmarks, MIMICEchoQA and SurgeryVideoQA. On these benchmarks, the 2B model achieves gains of 99.1% and 98.1%, while the 7B model shows gains of 22.5% and 52.1%, respectively, demonstrating the models' ability to generalize and perform biomedical video understanding on cleaner and more standardized datasets than those seen during training. These results suggest that educational videos created for human learning offer a surprisingly effective training signal for biomedical VLMs.
How well do LLMs cite relevant medical references? An evaluation framework and analyses
Feb 03, 2024Abstract:Large language models (LLMs) are currently being used to answer medical questions across a variety of clinical domains. Recent top-performing commercial LLMs, in particular, are also capable of citing sources to support their responses. In this paper, we ask: do the sources that LLMs generate actually support the claims that they make? To answer this, we propose three contributions. First, as expert medical annotations are an expensive and time-consuming bottleneck for scalable evaluation, we demonstrate that GPT-4 is highly accurate in validating source relevance, agreeing 88% of the time with a panel of medical doctors. Second, we develop an end-to-end, automated pipeline called \textit{SourceCheckup} and use it to evaluate five top-performing LLMs on a dataset of 1200 generated questions, totaling over 40K pairs of statements and sources. Interestingly, we find that between ~50% to 90% of LLM responses are not fully supported by the sources they provide. We also evaluate GPT-4 with retrieval augmented generation (RAG) and find that, even still, around 30\% of individual statements are unsupported, while nearly half of its responses are not fully supported. Third, we open-source our curated dataset of medical questions and expert annotations for future evaluations. Given the rapid pace of LLM development and the potential harms of incorrect or outdated medical information, it is crucial to also understand and quantify their capability to produce relevant, trustworthy medical references.
ChatGPT Exhibits Gender and Racial Biases in Acute Coronary Syndrome Management
Nov 10, 2023Abstract:Recent breakthroughs in large language models (LLMs) have led to their rapid dissemination and widespread use. One early application has been to medicine, where LLMs have been investigated to streamline clinical workflows and facilitate clinical analysis and decision-making. However, a leading barrier to the deployment of Artificial Intelligence (AI) and in particular LLMs has been concern for embedded gender and racial biases. Here, we evaluate whether a leading LLM, ChatGPT 3.5, exhibits gender and racial bias in clinical management of acute coronary syndrome (ACS). We find that specifying patients as female, African American, or Hispanic resulted in a decrease in guideline recommended medical management, diagnosis, and symptom management of ACS. Most notably, the largest disparities were seen in the recommendation of coronary angiography or stress testing for the diagnosis and further intervention of ACS and recommendation of high intensity statins. These disparities correlate with biases that have been observed clinically and have been implicated in the differential gender and racial morbidity and mortality outcomes of ACS and coronary artery disease. Furthermore, we find that the largest disparities are seen during unstable angina, where fewer explicit clinical guidelines exist. Finally, we find that through asking ChatGPT 3.5 to explain its reasoning prior to providing an answer, we are able to improve clinical accuracy and mitigate instances of gender and racial biases. This is among the first studies to demonstrate that the gender and racial biases that LLMs exhibit do in fact affect clinical management. Additionally, we demonstrate that existing strategies that improve LLM performance not only improve LLM performance in clinical management, but can also be used to mitigate gender and racial biases.
A computational geometry approach for modeling neuronal fiber pathways
Aug 02, 2021



Abstract:We propose a novel and efficient algorithm to model high-level topological structures of neuronal fibers. Tractography constructs complex neuronal fibers in three dimensions that exhibit the geometry of white matter pathways in the brain. However, most tractography analysis methods are time consuming and intractable. We develop a computational geometry-based tractography representation that aims to simplify the connectivity of white matter fibers. Given the trajectories of neuronal fiber pathways, we model the evolution of trajectories that encodes geometrically significant events and calculate their point correspondence in the 3D brain space. Trajectory inter-distance is used as a parameter to control the granularity of the model that allows local or global representation of the tractogram. Using diffusion MRI data from Alzheimer's patient study, we extract tractography features from our model for distinguishing the Alzheimer's subject from the normal control. Software implementation of our algorithm is available on GitHub.
Predicting Fluid Intelligence of Children using T1-weighted MR Images and a StackNet
May 07, 2019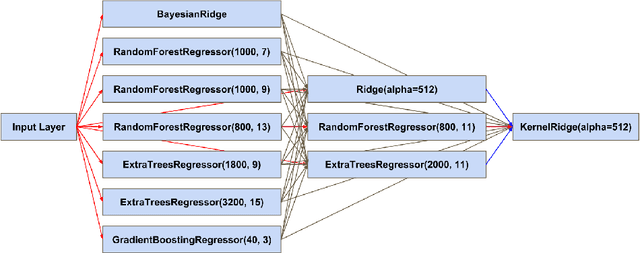

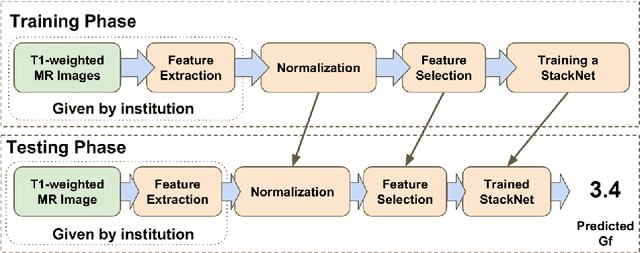
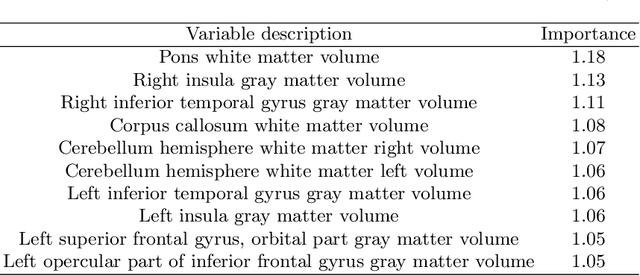
Abstract:In this work, we utilize T1-weighted MR images and StackNet to predict fluid intelligence in adolescents. Our framework includes feature extraction, feature normalization, feature denoising, feature selection, training a StackNet, and predicting fluid intelligence. The extracted feature is the distribution of different brain tissues in different brain parcellation regions. The proposed StackNet consists of three layers and 11 models. Each layer uses the predictions from all previous layers including the input layer. The proposed StackNet is tested on a public benchmark Adolescent Brain Cognitive Development Neurocognitive Prediction Challenge 2019 and achieves a mean squared error of 82.42 on the combined training and validation set with 10-fold cross-validation. In addition, the proposed StackNet also achieves a mean squared error of 94.25 on the testing data. The source code is available on GitHub.
Fully Automated Volumetric Classification in CT Scans for Diagnosis and Analysis of Normal Pressure Hydrocephalus
Jan 25, 2019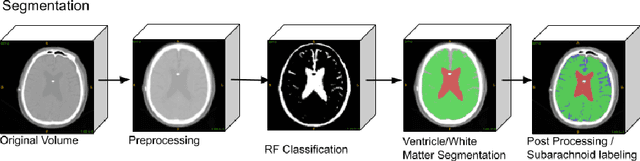
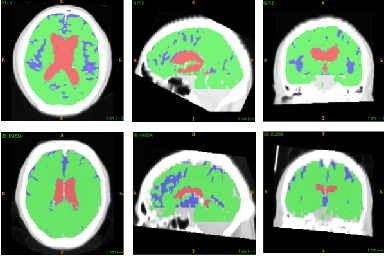
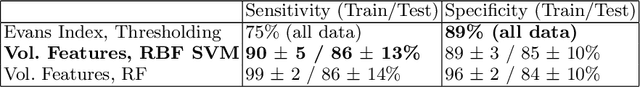
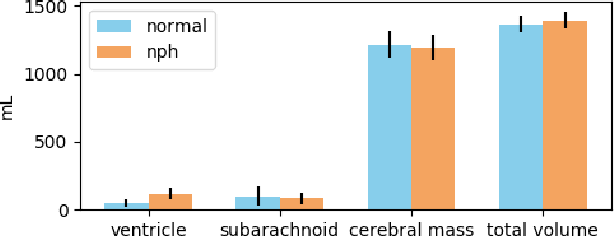
Abstract:Normal Pressure Hydrocephalus (NPH) is one of the few reversible forms of dementia. Due to their low cost and versatility, Computed Tomography (CT) scans have long been used as an aid to help diagnose intracerebral anomalies such as NPH. However, because CT imaging presents 2-dimensional slices of a 3-dimensional volume, recapitulating the ventricular space in 3-dimensions to facilitate the diagnosis of NPH poses numerous challenges such as head rotation and human error. As such, no well-defined and effective protocol currently exists for the analysis of CT scan-based ventricular, white matter and subarachnoid space volumes in the setting of NPH. The Evan's ratio, an approximation of the ratio of ventricle to brain volume using only one 2D slice of the scan, has been proposed but is not robust. Instead of manually measuring a 2-dimensional proxy for the ratio of ventricle volume to brain volume, this study proposes an automated method of calculating the brain volumes for better recognition of NPH from a radiological standpoint. The method first aligns the subject CT volume to a common space through an affine transformation, then uses a random forest classifier to mask relevant tissue types. A 3D morphological segmentation method is used to partition the brain volume, which in turn is used to train machine learning methods to classify the subjects into non-NPH vs. NPH based on volumetric information.
Brain Tumor Segmentation and Tractographic Feature Extraction from Structural MR Images for Overall Survival Prediction
Oct 10, 2018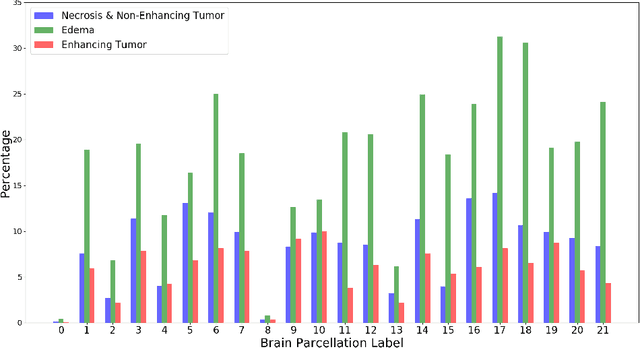
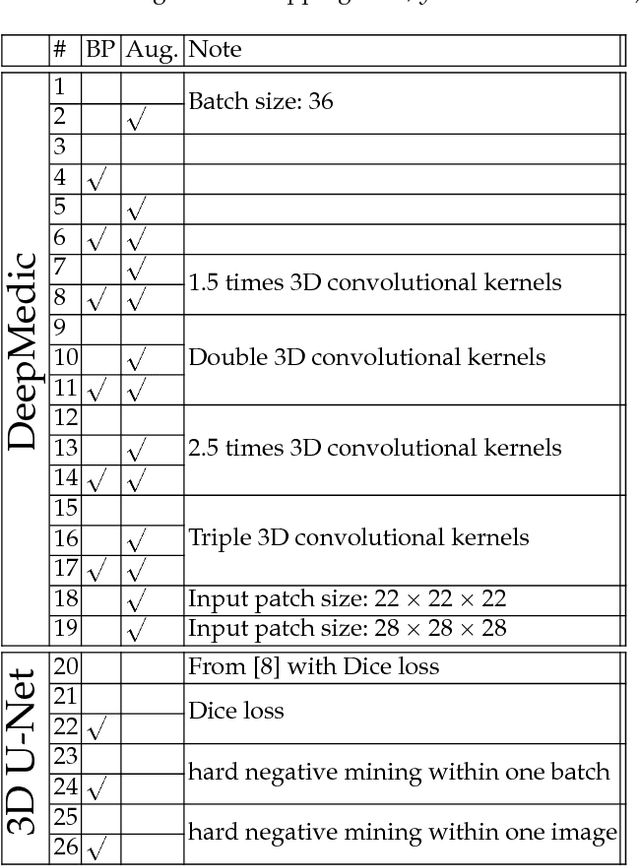
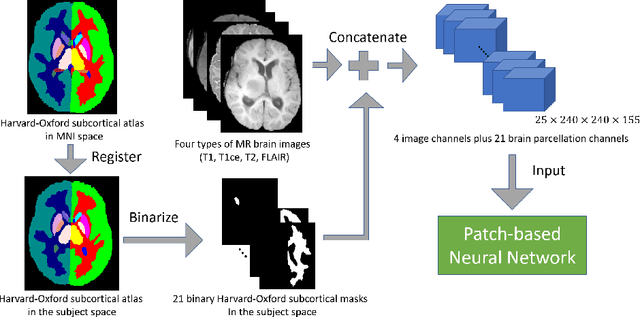

Abstract:This paper introduces a novel methodology to integrate human brain connectomics and parcellation for brain tumor segmentation and survival prediction. For segmentation, we utilize an existing brain parcellation atlas in the MNI152 1mm space and map this parcellation to each individual subject data. We use deep neural network architectures together with hard negative mining to achieve the final voxel level classification. For survival prediction, we present a new method for combining features from connectomics data, brain parcellation information, and the brain tumor mask. We leverage the average connectome information from the Human Connectome Project and map each subject brain volume onto this common connectome space. From this, we compute tractographic features that describe potential neural disruptions due to the brain tumor. These features are then used to predict the overall survival of the subjects. The main novelty in the proposed methods is the use of normalized brain parcellation data and tractography data from the human connectome project for analyzing MR images for segmentation and survival prediction. Experimental results are reported on the BraTS2018 data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge