S. Shailja
Deep Learning Enabled Time-Lapse 3D Cell Analysis
Aug 17, 2022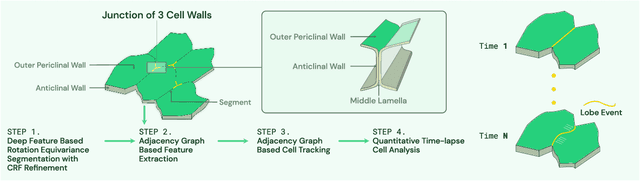
Abstract:This paper presents a method for time-lapse 3D cell analysis. Specifically, we consider the problem of accurately localizing and quantitatively analyzing sub-cellular features, and for tracking individual cells from time-lapse 3D confocal cell image stacks. The heterogeneity of cells and the volume of multi-dimensional images presents a major challenge for fully automated analysis of morphogenesis and development of cells. This paper is motivated by the pavement cell growth process, and building a quantitative morphogenesis model. We propose a deep feature based segmentation method to accurately detect and label each cell region. An adjacency graph based method is used to extract sub-cellular features of the segmented cells. Finally, the robust graph based tracking algorithm using multiple cell features is proposed for associating cells at different time instances. Extensive experiment results are provided and demonstrate the robustness of the proposed method. The code is available on Github and the method is available as a service through the BisQue portal.
A computational geometry approach for modeling neuronal fiber pathways
Aug 02, 2021



Abstract:We propose a novel and efficient algorithm to model high-level topological structures of neuronal fibers. Tractography constructs complex neuronal fibers in three dimensions that exhibit the geometry of white matter pathways in the brain. However, most tractography analysis methods are time consuming and intractable. We develop a computational geometry-based tractography representation that aims to simplify the connectivity of white matter fibers. Given the trajectories of neuronal fiber pathways, we model the evolution of trajectories that encodes geometrically significant events and calculate their point correspondence in the 3D brain space. Trajectory inter-distance is used as a parameter to control the granularity of the model that allows local or global representation of the tractogram. Using diffusion MRI data from Alzheimer's patient study, we extract tractography features from our model for distinguishing the Alzheimer's subject from the normal control. Software implementation of our algorithm is available on GitHub.
Semi supervised segmentation and graph-based tracking of 3D nuclei in time-lapse microscopy
Oct 26, 2020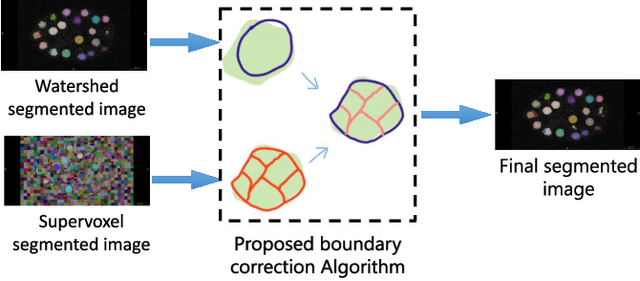
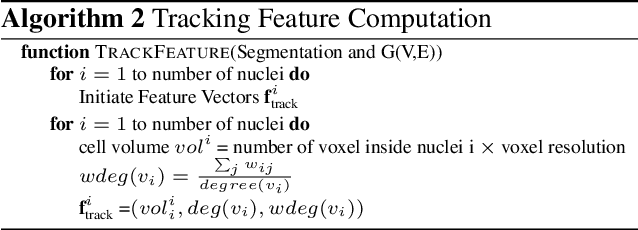
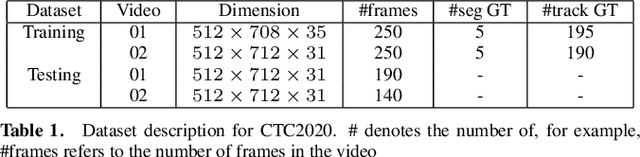
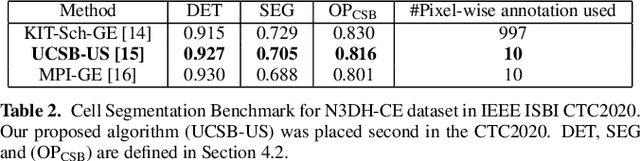
Abstract:We propose a novel weakly supervised method to improve the boundary of the 3D segmented nuclei utilizing an over-segmented image. This is motivated by the observation that current state-of-the-art deep learning methods do not result in accurate boundaries when the training data is weakly annotated. Towards this, a 3D U-Net is trained to get the centroid of the nuclei and integrated with a simple linear iterative clustering (SLIC) supervoxel algorithm that provides better adherence to cluster boundaries. To track these segmented nuclei, our algorithm utilizes the relative nuclei location depicting the processes of nuclei division and apoptosis. The proposed algorithmic pipeline achieves better segmentation performance compared to the state-of-the-art method in Cell Tracking Challenge (CTC) 2019 and comparable performance to state-of-the-art methods in IEEE ISBI CTC2020 while utilizing very few pixel-wise annotated data. Detailed experimental results are provided, and the source code is available on GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge