Semi supervised segmentation and graph-based tracking of 3D nuclei in time-lapse microscopy
Paper and Code
Oct 26, 2020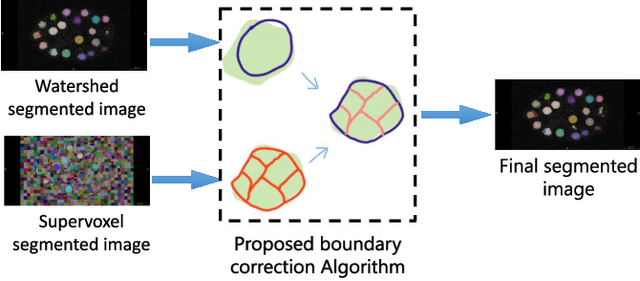
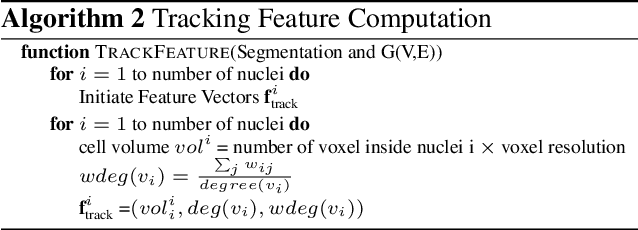
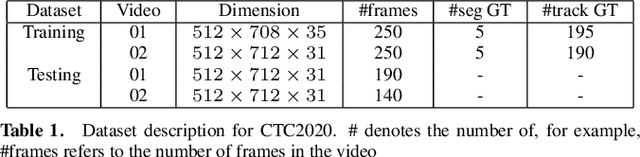
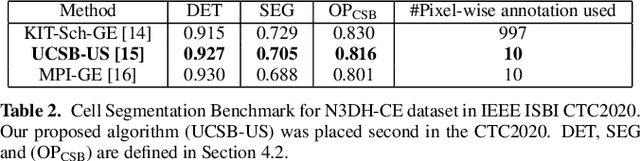
We propose a novel weakly supervised method to improve the boundary of the 3D segmented nuclei utilizing an over-segmented image. This is motivated by the observation that current state-of-the-art deep learning methods do not result in accurate boundaries when the training data is weakly annotated. Towards this, a 3D U-Net is trained to get the centroid of the nuclei and integrated with a simple linear iterative clustering (SLIC) supervoxel algorithm that provides better adherence to cluster boundaries. To track these segmented nuclei, our algorithm utilizes the relative nuclei location depicting the processes of nuclei division and apoptosis. The proposed algorithmic pipeline achieves better segmentation performance compared to the state-of-the-art method in Cell Tracking Challenge (CTC) 2019 and comparable performance to state-of-the-art methods in IEEE ISBI CTC2020 while utilizing very few pixel-wise annotated data. Detailed experimental results are provided, and the source code is available on GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge