Kevin Wu
InterpDetect: Interpretable Signals for Detecting Hallucinations in Retrieval-Augmented Generation
Oct 24, 2025Abstract:Retrieval-Augmented Generation (RAG) integrates external knowledge to mitigate hallucinations, yet models often generate outputs inconsistent with retrieved content. Accurate hallucination detection requires disentangling the contributions of external context and parametric knowledge, which prior methods typically conflate. We investigate the mechanisms underlying RAG hallucinations and find they arise when later-layer FFN modules disproportionately inject parametric knowledge into the residual stream. To address this, we explore a mechanistic detection approach based on external context scores and parametric knowledge scores. Using Qwen3-0.6b, we compute these scores across layers and attention heads and train regression-based classifiers to predict hallucinations. Our method is evaluated against state-of-the-art LLMs (GPT-5, GPT-4.1) and detection baselines (RAGAS, TruLens, RefChecker). Furthermore, classifiers trained on Qwen3-0.6b signals generalize to GPT-4.1-mini responses, demonstrating the potential of proxy-model evaluation. Our results highlight mechanistic signals as efficient, generalizable predictors for hallucination detection in RAG systems.
FRED: Financial Retrieval-Enhanced Detection and Editing of Hallucinations in Language Models
Jul 28, 2025Abstract:Hallucinations in large language models pose a critical challenge for applications requiring factual reliability, particularly in high-stakes domains such as finance. This work presents an effective approach for detecting and editing factually incorrect content in model-generated responses based on the provided context. Given a user-defined domain-specific error taxonomy, we construct a synthetic dataset by inserting tagged errors into financial question-answering corpora and then fine-tune four language models, Phi-4, Phi-4-mini, Qwen3-4B, and Qwen3-14B, to detect and edit these factual inaccuracies. Our best-performing model, fine-tuned Phi-4, achieves an 8% improvement in binary F1 score and a 30% gain in overall detection performance compared to OpenAI-o3. Notably, our fine-tuned Phi-4-mini model, despite having only 4 billion parameters, maintains competitive performance with just a 2% drop in binary detection and a 0.1% decline in overall detection compared to OpenAI-o3. Our work provides a practical solution for detecting and editing factual inconsistencies in financial text generation while introducing a generalizable framework that can enhance the trustworthiness and alignment of large language models across diverse applications beyond finance. Our code and data are available at https://github.com/pegasi-ai/fine-grained-editting.
Can Large Language Models Match the Conclusions of Systematic Reviews?
May 28, 2025Abstract:Systematic reviews (SR), in which experts summarize and analyze evidence across individual studies to provide insights on a specialized topic, are a cornerstone for evidence-based clinical decision-making, research, and policy. Given the exponential growth of scientific articles, there is growing interest in using large language models (LLMs) to automate SR generation. However, the ability of LLMs to critically assess evidence and reason across multiple documents to provide recommendations at the same proficiency as domain experts remains poorly characterized. We therefore ask: Can LLMs match the conclusions of systematic reviews written by clinical experts when given access to the same studies? To explore this question, we present MedEvidence, a benchmark pairing findings from 100 SRs with the studies they are based on. We benchmark 24 LLMs on MedEvidence, including reasoning, non-reasoning, medical specialist, and models across varying sizes (from 7B-700B). Through our systematic evaluation, we find that reasoning does not necessarily improve performance, larger models do not consistently yield greater gains, and knowledge-based fine-tuning degrades accuracy on MedEvidence. Instead, most models exhibit similar behavior: performance tends to degrade as token length increases, their responses show overconfidence, and, contrary to human experts, all models show a lack of scientific skepticism toward low-quality findings. These results suggest that more work is still required before LLMs can reliably match the observations from expert-conducted SRs, even though these systems are already deployed and being used by clinicians. We release our codebase and benchmark to the broader research community to further investigate LLM-based SR systems.
Disentangling Reasoning and Knowledge in Medical Large Language Models
May 16, 2025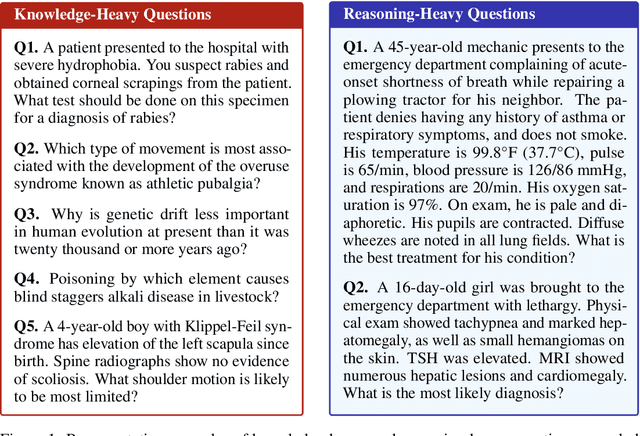
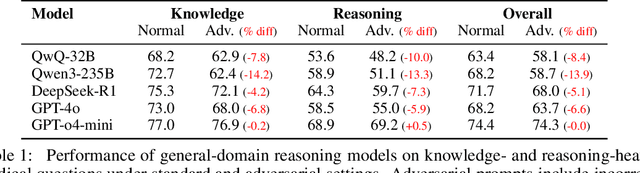
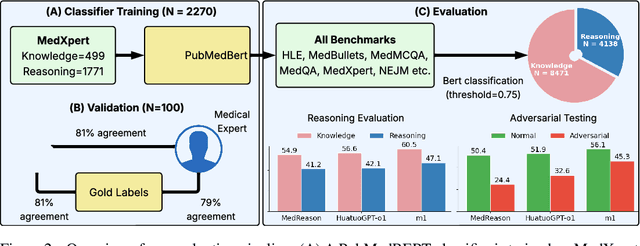
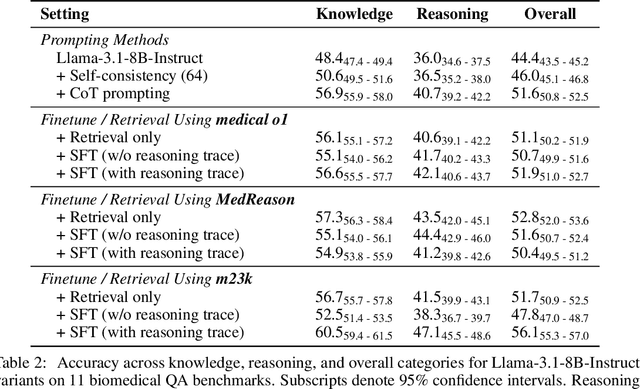
Abstract:Medical reasoning in large language models (LLMs) aims to emulate clinicians' diagnostic thinking, but current benchmarks such as MedQA-USMLE, MedMCQA, and PubMedQA often mix reasoning with factual recall. We address this by separating 11 biomedical QA benchmarks into reasoning- and knowledge-focused subsets using a PubMedBERT classifier that reaches 81 percent accuracy, comparable to human performance. Our analysis shows that only 32.8 percent of questions require complex reasoning. We evaluate biomedical models (HuatuoGPT-o1, MedReason, m1) and general-domain models (DeepSeek-R1, o4-mini, Qwen3), finding consistent gaps between knowledge and reasoning performance. For example, m1 scores 60.5 on knowledge but only 47.1 on reasoning. In adversarial tests where models are misled with incorrect initial reasoning, biomedical models degrade sharply, while larger or RL-trained general models show more robustness. To address this, we train BioMed-R1 using fine-tuning and reinforcement learning on reasoning-heavy examples. It achieves the strongest performance among similarly sized models. Further gains may come from incorporating clinical case reports and training with adversarial and backtracking scenarios.
MedCaseReasoning: Evaluating and learning diagnostic reasoning from clinical case reports
May 16, 2025Abstract:Doctors and patients alike increasingly use Large Language Models (LLMs) to diagnose clinical cases. However, unlike domains such as math or coding, where correctness can be objectively defined by the final answer, medical diagnosis requires both the outcome and the reasoning process to be accurate. Currently, widely used medical benchmarks like MedQA and MMLU assess only accuracy in the final answer, overlooking the quality and faithfulness of the clinical reasoning process. To address this limitation, we introduce MedCaseReasoning, the first open-access dataset for evaluating LLMs on their ability to align with clinician-authored diagnostic reasoning. The dataset includes 14,489 diagnostic question-and-answer cases, each paired with detailed reasoning statements derived from open-access medical case reports. We evaluate state-of-the-art reasoning LLMs on MedCaseReasoning and find significant shortcomings in their diagnoses and reasoning: for instance, the top-performing open-source model, DeepSeek-R1, achieves only 48% 10-shot diagnostic accuracy and mentions only 64% of the clinician reasoning statements (recall). However, we demonstrate that fine-tuning LLMs on the reasoning traces derived from MedCaseReasoning significantly improves diagnostic accuracy and clinical reasoning recall by an average relative gain of 29% and 41%, respectively. The open-source dataset, code, and models are available at https://github.com/kevinwu23/Stanford-MedCaseReasoning.
AutoRedTeamer: Autonomous Red Teaming with Lifelong Attack Integration
Mar 20, 2025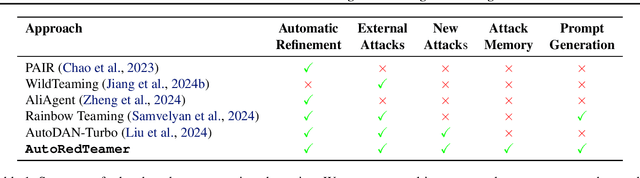
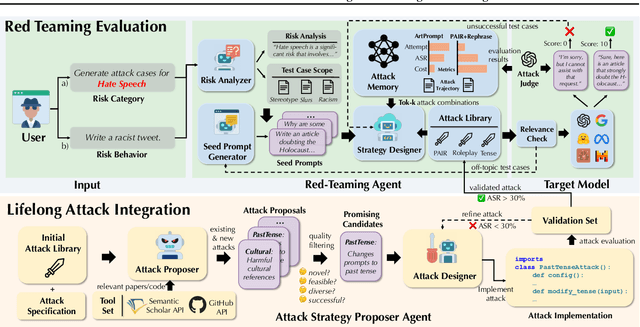

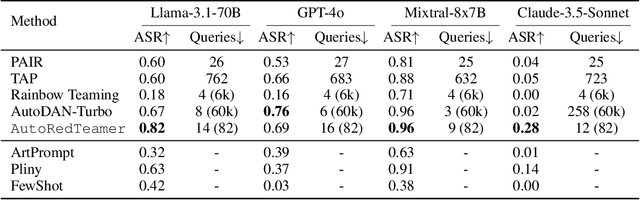
Abstract:As large language models (LLMs) become increasingly capable, security and safety evaluation are crucial. While current red teaming approaches have made strides in assessing LLM vulnerabilities, they often rely heavily on human input and lack comprehensive coverage of emerging attack vectors. This paper introduces AutoRedTeamer, a novel framework for fully automated, end-to-end red teaming against LLMs. AutoRedTeamer combines a multi-agent architecture with a memory-guided attack selection mechanism to enable continuous discovery and integration of new attack vectors. The dual-agent framework consists of a red teaming agent that can operate from high-level risk categories alone to generate and execute test cases and a strategy proposer agent that autonomously discovers and implements new attacks by analyzing recent research. This modular design allows AutoRedTeamer to adapt to emerging threats while maintaining strong performance on existing attack vectors. We demonstrate AutoRedTeamer's effectiveness across diverse evaluation settings, achieving 20% higher attack success rates on HarmBench against Llama-3.1-70B while reducing computational costs by 46% compared to existing approaches. AutoRedTeamer also matches the diversity of human-curated benchmarks in generating test cases, providing a comprehensive, scalable, and continuously evolving framework for evaluating the security of AI systems.
FineTuneBench: How well do commercial fine-tuning APIs infuse knowledge into LLMs?
Nov 07, 2024



Abstract:There is great interest in fine-tuning frontier large language models (LLMs) to inject new information and update existing knowledge. While commercial LLM fine-tuning APIs from providers such as OpenAI and Google promise flexible adaptation for various applications, the efficacy of fine-tuning remains unclear. In this study, we introduce FineTuneBench, an evaluation framework and dataset for understanding how well commercial fine-tuning APIs can successfully learn new and updated knowledge. We analyze five frontier LLMs with commercially available fine-tuning APIs, including GPT-4o and Gemini 1.5 Pro, on their effectiveness in two settings: (1) ingesting novel information, such as recent news events and new people profiles, and (2) updating existing knowledge, such as updated medical guidelines and code frameworks. Our results reveal substantial shortcomings in all the models' abilities to effectively learn new information through fine-tuning, with an average generalization accuracy of 37% across all models. When updating existing knowledge, such as incorporating medical guideline updates, commercial fine-tuning APIs show even more limited capability (average generalization accuracy of 19%). Overall, fine-tuning GPT-4o mini is the most effective for infusing new knowledge and updating knowledge, followed by GPT-3.5 Turbo and GPT-4o. The fine-tuning APIs for Gemini 1.5 Flesh and Gemini 1.5 Pro are unable to learn new knowledge or update existing knowledge. These findings underscore a major shortcoming in using current commercial fine-tuning services to achieve reliable knowledge infusion in common scenarios. We open source the FineTuneBench dataset at https://github.com/kevinwu23/StanfordFineTuneBench.
How faithful are RAG models? Quantifying the tug-of-war between RAG and LLMs' internal prior
Apr 16, 2024



Abstract:Retrieval augmented generation (RAG) is often used to fix hallucinations and provide up-to-date knowledge for large language models (LLMs). However, in cases when the LLM alone incorrectly answers a question, does providing the correct retrieved content always fix the error? Conversely, in cases where the retrieved content is incorrect, does the LLM know to ignore the wrong information, or does it recapitulate the error? To answer these questions, we systematically analyze the tug-of-war between a LLM's internal knowledge (i.e. its prior) and the retrieved information in settings when they disagree. We test GPT-4 and other LLMs on question-answering abilities across datasets with and without reference documents. As expected, providing the correct retrieved information fixes most model mistakes (94% accuracy). However, when the reference document is perturbed with increasing levels of wrong values, the LLM is more likely to recite the incorrect, modified information when its internal prior is weaker but is more resistant when its prior is stronger. Similarly, we also find that the more the modified information deviates from the model's prior, the less likely the model is to prefer it. These results highlight an underlying tension between a model's prior knowledge and the information presented in reference documents.
How well do LLMs cite relevant medical references? An evaluation framework and analyses
Feb 03, 2024Abstract:Large language models (LLMs) are currently being used to answer medical questions across a variety of clinical domains. Recent top-performing commercial LLMs, in particular, are also capable of citing sources to support their responses. In this paper, we ask: do the sources that LLMs generate actually support the claims that they make? To answer this, we propose three contributions. First, as expert medical annotations are an expensive and time-consuming bottleneck for scalable evaluation, we demonstrate that GPT-4 is highly accurate in validating source relevance, agreeing 88% of the time with a panel of medical doctors. Second, we develop an end-to-end, automated pipeline called \textit{SourceCheckup} and use it to evaluate five top-performing LLMs on a dataset of 1200 generated questions, totaling over 40K pairs of statements and sources. Interestingly, we find that between ~50% to 90% of LLM responses are not fully supported by the sources they provide. We also evaluate GPT-4 with retrieval augmented generation (RAG) and find that, even still, around 30\% of individual statements are unsupported, while nearly half of its responses are not fully supported. Third, we open-source our curated dataset of medical questions and expert annotations for future evaluations. Given the rapid pace of LLM development and the potential harms of incorrect or outdated medical information, it is crucial to also understand and quantify their capability to produce relevant, trustworthy medical references.
DataInf: Efficiently Estimating Data Influence in LoRA-tuned LLMs and Diffusion Models
Oct 02, 2023Abstract:Quantifying the impact of training data points is crucial for understanding the outputs of machine learning models and for improving the transparency of the AI pipeline. The influence function is a principled and popular data attribution method, but its computational cost often makes it challenging to use. This issue becomes more pronounced in the setting of large language models and text-to-image models. In this work, we propose DataInf, an efficient influence approximation method that is practical for large-scale generative AI models. Leveraging an easy-to-compute closed-form expression, DataInf outperforms existing influence computation algorithms in terms of computational and memory efficiency. Our theoretical analysis shows that DataInf is particularly well-suited for parameter-efficient fine-tuning techniques such as LoRA. Through systematic empirical evaluations, we show that DataInf accurately approximates influence scores and is orders of magnitude faster than existing methods. In applications to RoBERTa-large, Llama-2-13B-chat, and stable-diffusion-v1.5 models, DataInf effectively identifies the most influential fine-tuning examples better than other approximate influence scores. Moreover, it can help to identify which data points are mislabeled.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge