Emmanuel Mignot
Stanford Sleep Bench: Evaluating Polysomnography Pre-training Methods for Sleep Foundation Models
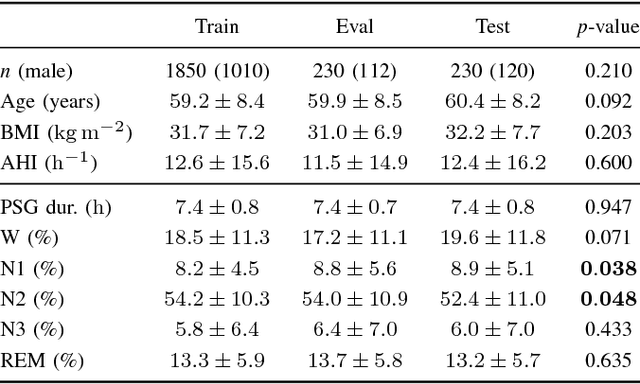
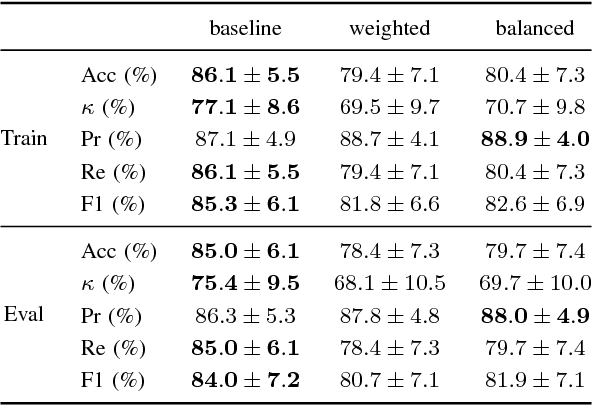
Dec 10, 2025Abstract:Polysomnography (PSG), the gold standard test for sleep analysis, generates vast amounts of multimodal clinical data, presenting an opportunity to leverage self-supervised representation learning (SSRL) for pre-training foundation models to enhance sleep analysis. However, progress in sleep foundation models is hindered by two key limitations: (1) the lack of a shared dataset and benchmark with diverse tasks for training and evaluation, and (2) the absence of a systematic evaluation of SSRL approaches across sleep-related tasks. To address these gaps, we introduce Stanford Sleep Bench, a large-scale PSG dataset comprising 17,467 recordings totaling over 163,000 hours from a major sleep clinic, including 13 clinical disease prediction tasks alongside canonical sleep-related tasks such as sleep staging, apnea diagnosis, and age estimation. We systematically evaluate SSRL pre-training methods on Stanford Sleep Bench, assessing downstream performance across four tasks: sleep staging, apnea diagnosis, age estimation, and disease and mortality prediction. Our results show that multiple pretraining methods achieve comparable performance for sleep staging, apnea diagnosis, and age estimation. However, for mortality and disease prediction, contrastive learning significantly outperforms other approaches while also converging faster during pretraining. To facilitate reproducibility and advance sleep research, we will release Stanford Sleep Bench along with pretrained model weights, training pipelines, and evaluation code.
SleepFM: Multi-modal Representation Learning for Sleep Across Brain Activity, ECG and Respiratory Signals
May 28, 2024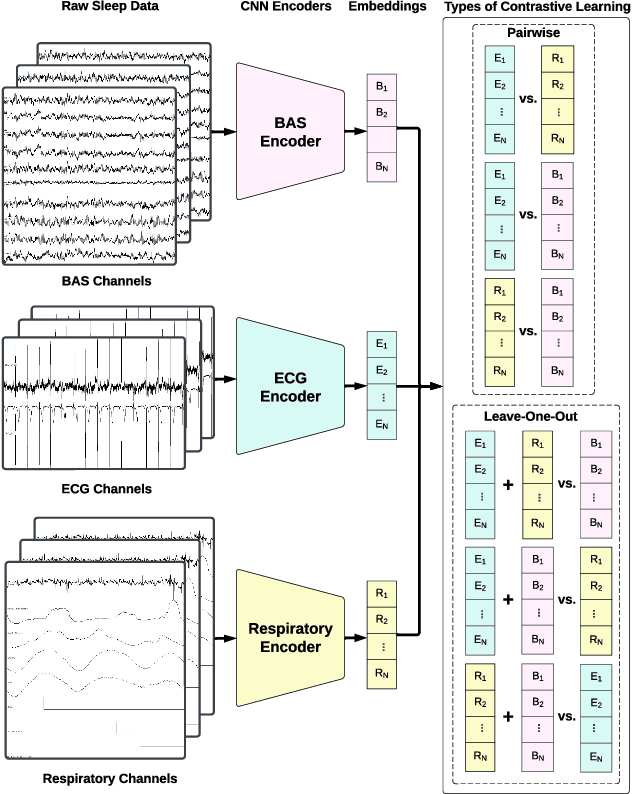
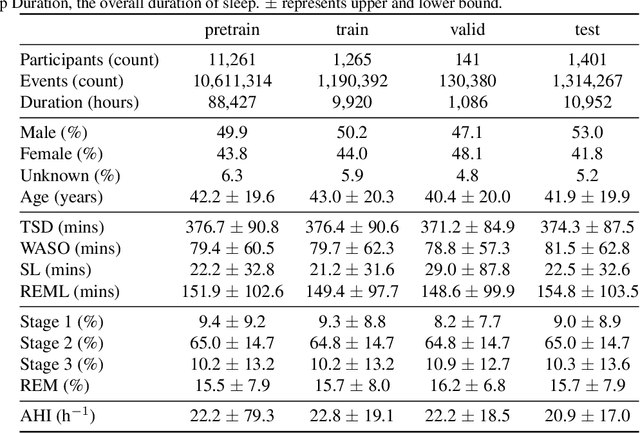
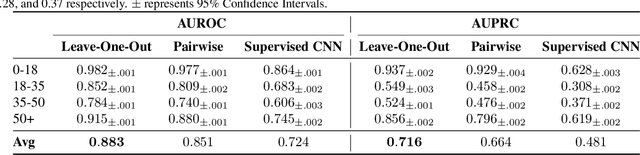
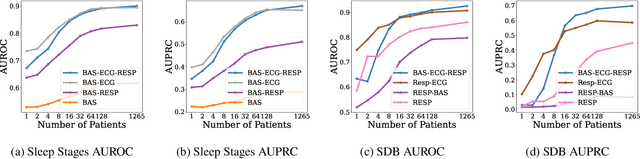
Abstract:Sleep is a complex physiological process evaluated through various modalities recording electrical brain, cardiac, and respiratory activities. We curate a large polysomnography dataset from over 14,000 participants comprising over 100,000 hours of multi-modal sleep recordings. Leveraging this extensive dataset, we developed SleepFM, the first multi-modal foundation model for sleep analysis. We show that a novel leave-one-out approach for contrastive learning significantly improves downstream task performance compared to representations from standard pairwise contrastive learning. A logistic regression model trained on SleepFM's learned embeddings outperforms an end-to-end trained convolutional neural network (CNN) on sleep stage classification (macro AUROC 0.88 vs 0.72 and macro AUPRC 0.72 vs 0.48) and sleep disordered breathing detection (AUROC 0.85 vs 0.69 and AUPRC 0.77 vs 0.61). Notably, the learned embeddings achieve 48% top-1 average accuracy in retrieving the corresponding recording clips of other modalities from 90,000 candidates. This work demonstrates the value of holistic multi-modal sleep modeling to fully capture the richness of sleep recordings. SleepFM is open source and available at https://github.com/rthapa84/sleepfm-codebase.
RRWaveNet: A Compact End-to-End Multi-Scale Residual CNN for Robust PPG Respiratory Rate Estimation
Aug 18, 2022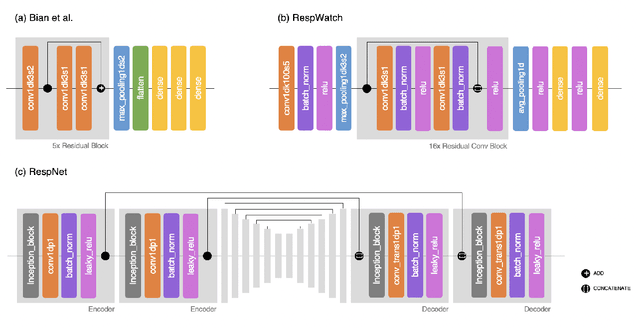
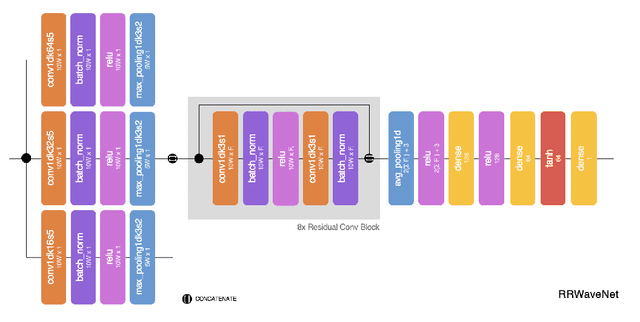
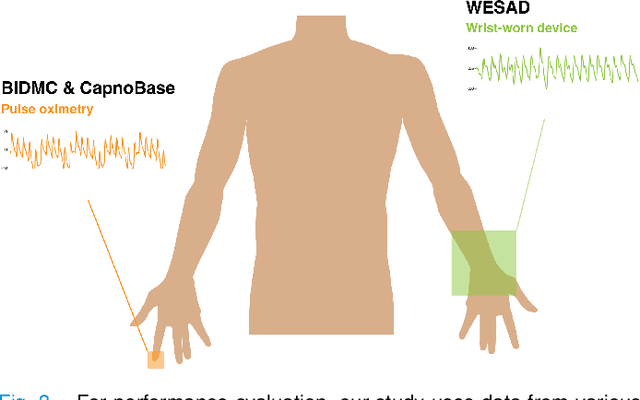
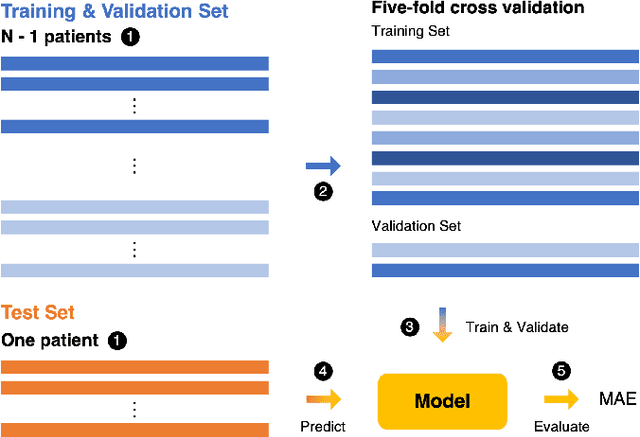
Abstract:Respiratory rate (RR) is an important biomarker as RR changes can reflect severe medical events such as heart disease, lung disease, and sleep disorders. Unfortunately, however, standard manual RR counting is prone to human error and cannot be performed continuously. This study proposes a method for continuously estimating RR, RRWaveNet. The method is a compact end-to-end deep learning model which does not require feature engineering and can use low-cost raw photoplethysmography (PPG) as input signal. RRWaveNet was tested subject-independently and compared to baseline in three datasets (BIDMC, CapnoBase, and WESAD) and using three window sizes (16, 32, and 64 seconds). RRWaveNet outperformed current state-of-the-art methods with mean absolute errors at optimal window size of 1.66 \pm 1.01, 1.59 \pm 1.08, and 1.92 \pm 0.96 breaths per minute for each dataset. In remote monitoring settings, such as in the WESAD dataset, we apply transfer learning to two other ICU datasets, reducing the MAE to 1.52 \pm 0.50 breaths per minute, showing this model allows accurate and practical estimation of RR on affordable and wearable devices. Our study shows feasibility of remote RR monitoring in the context of telemedicine and at home.
MSED: a multi-modal sleep event detection model for clinical sleep analysis
Jan 07, 2021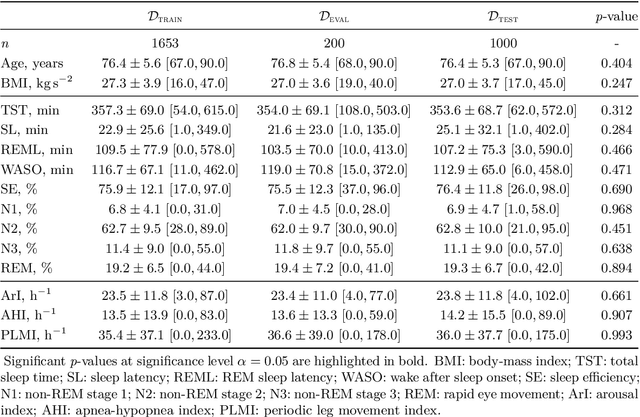
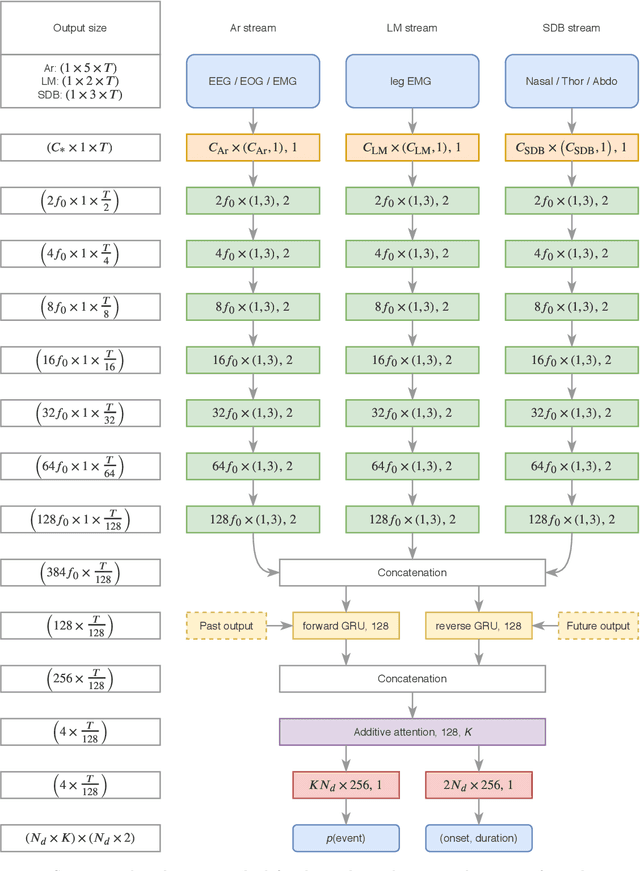
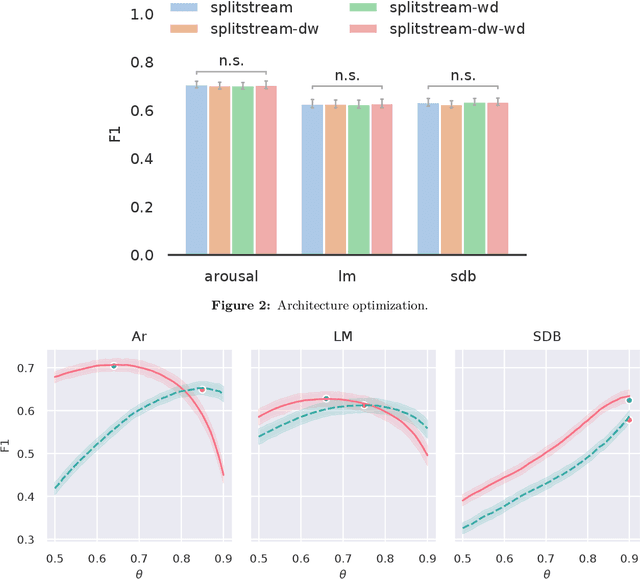
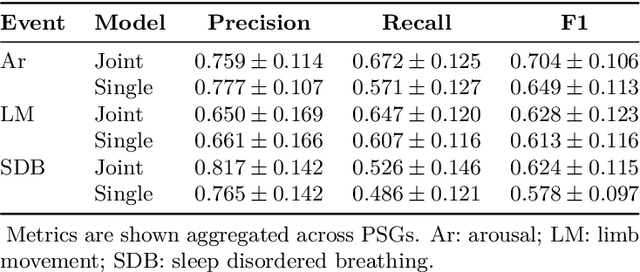
Abstract:Study objective: Clinical sleep analysis require manual analysis of sleep patterns for correct diagnosis of sleep disorders. Several studies show significant variability in scoring discrete sleep events. We wished to investigate, whether an automatic method could be used for detection of arousals (Ar), leg movements (LM) and sleep disordered breathing (SDB) events, and if the joint detection of these events performed better than having three separate models. Methods: We designed a single deep neural network architecture to jointly detect sleep events in a polysomnogram. We trained the model on 1653 recordings of individuals, and tested the optimized model on 1000 separate recordings. The performance of the model was quantified by F1, precision, and recall scores, and by correlating index values to clinical values using Pearson's correlation coefficient. Results: F1 scores for the optimized model was 0.70, 0.63, and 0.62 for Ar, LM, and SDB, respectively. The performance was higher, when detecting events jointly compared to corresponding single-event models. Index values computed from detected events correlated well with manual annotations ($r^2$ = 0.73, $r^2$ = 0.77, $r^2$ = 0.78, respectively). Conclusion: Detecting arousals, leg movements and sleep disordered breathing events jointly is possible, and the computed index values correlates well with human annotations.
Automatic sleep stage classification with deep residual networks in a mixed-cohort setting
Aug 21, 2020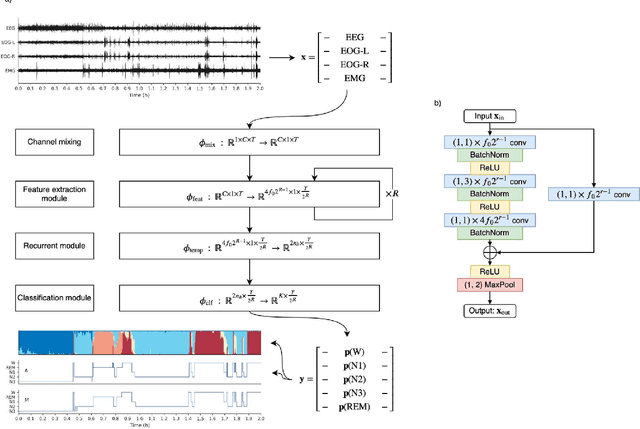
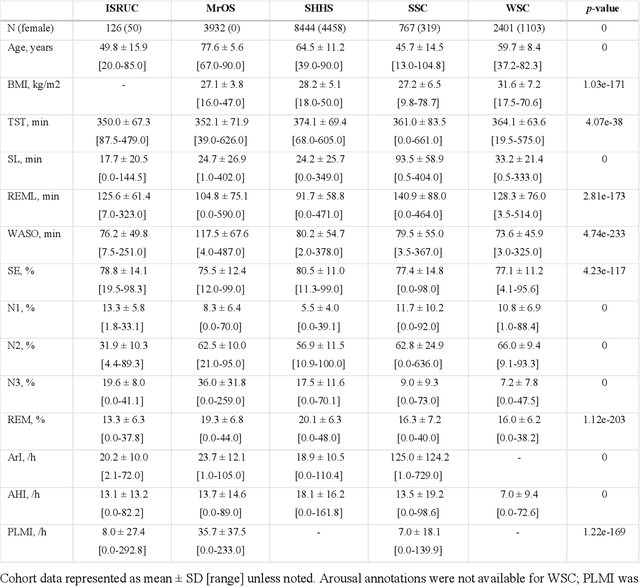
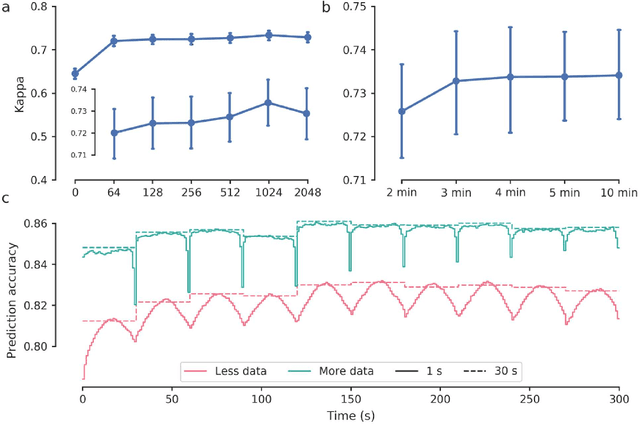
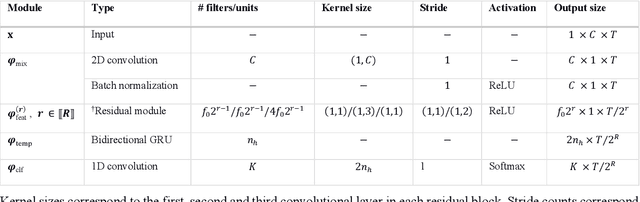
Abstract:Study Objectives: Sleep stage scoring is performed manually by sleep experts and is prone to subjective interpretation of scoring rules with low intra- and interscorer reliability. Many automatic systems rely on few small-scale databases for developing models, and generalizability to new datasets is thus unknown. We investigated a novel deep neural network to assess the generalizability of several large-scale cohorts. Methods: A deep neural network model was developed using 15684 polysomnography studies from five different cohorts. We applied four different scenarios: 1) impact of varying time-scales in the model; 2) performance of a single cohort on other cohorts of smaller, greater or equal size relative to the performance of other cohorts on a single cohort; 3) varying the fraction of mixed-cohort training data compared to using single-origin data; and 4) comparing models trained on combinations of data from 2, 3, and 4 cohorts. Results: Overall classification accuracy improved with increasing fractions of training data (0.25$\%$: 0.782 $\pm$ 0.097, 95$\%$ CI [0.777-0.787]; 100$\%$: 0.869 $\pm$ 0.064, 95$\%$ CI [0.864-0.872]), and with increasing number of data sources (2: 0.788 $\pm$ 0.102, 95$\%$ CI [0.787-0.790]; 3: 0.808 $\pm$ 0.092, 95$\%$ CI [0.807-0.810]; 4: 0.821 $\pm$ 0.085, 95$\%$ CI [0.819-0.823]). Different cohorts show varying levels of generalization to other cohorts. Conclusions: Automatic sleep stage scoring systems based on deep learning algorithms should consider as much data as possible from as many sources available to ensure proper generalization. Public datasets for benchmarking should be made available for future research.
Deep transfer learning for improving single-EEG arousal detection
May 07, 2020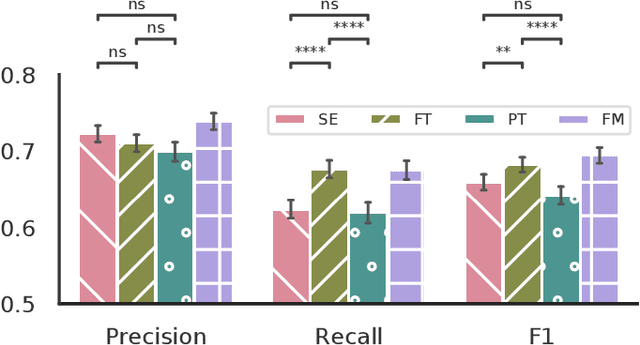
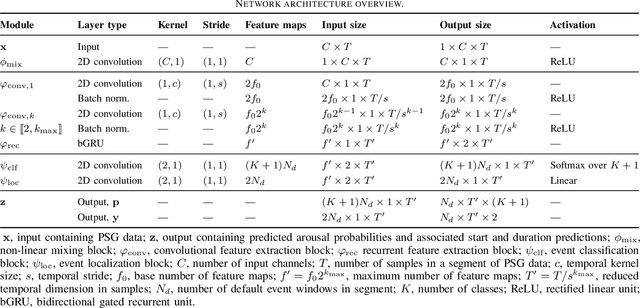
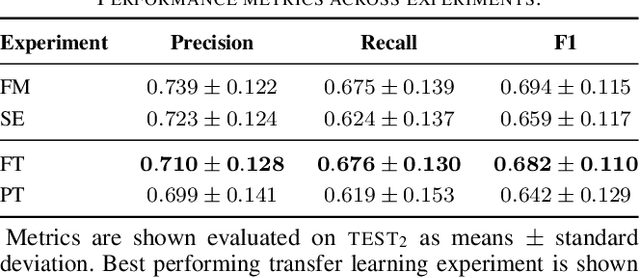
Abstract:Datasets in sleep science present challenges for machine learning algorithms due to differences in recording setups across clinics. We investigate two deep transfer learning strategies for overcoming the channel mismatch problem for cases where two datasets do not contain exactly the same setup leading to degraded performance in single-EEG models. Specifically, we train a baseline model on multivariate polysomnography data and subsequently replace the first two layers to prepare the architecture for single-channel electroencephalography data. Using a fine-tuning strategy, our model yields similar performance to the baseline model (F1=0.682 and F1=0.694, respectively), and was significantly better than a comparable single-channel model. Our results are promising for researchers working with small databases who wish to use deep learning models pre-trained on larger databases.
Towards a Flexible Deep Learning Method for Automatic Detection of Clinically Relevant Multi-Modal Events in the Polysomnogram
May 16, 2019



Abstract:Much attention has been given to automatic sleep staging algorithms in past years, but the detection of discrete events in sleep studies is also crucial for precise characterization of sleep patterns and possible diagnosis of sleep disorders. We propose here a deep learning model for automatic detection and annotation of arousals and leg movements. Both of these are commonly seen during normal sleep, while an excessive amount of either is linked to disrupted sleep patterns, excessive daytime sleepiness impacting quality of life, and various sleep disorders. Our model was trained on 1,485 subjects and tested on 1,000 separate recordings of sleep. We tested two different experimental setups and found optimal arousal detection was attained by including a recurrent neural network module in our default model with a dynamic default event window (F1 = 0.75), while optimal leg movement detection was attained using a static event window (F1 = 0.65). Our work show promise while still allowing for improvements. Specifically, future research will explore the proposed model as a general-purpose sleep analysis model.
DOSED: a deep learning approach to detect multiple sleep micro-events in EEG signal
Dec 07, 2018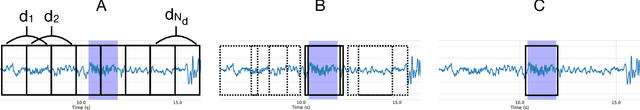
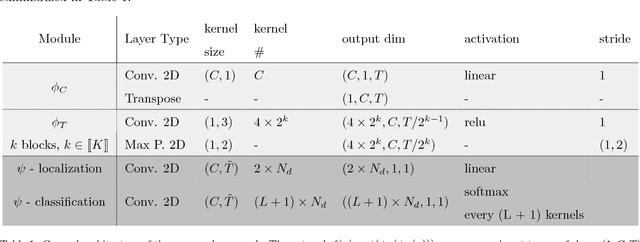
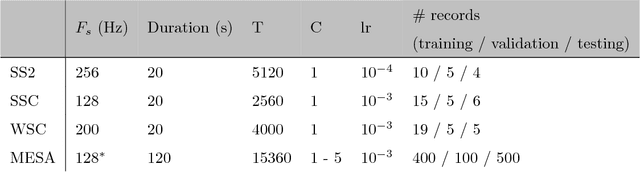
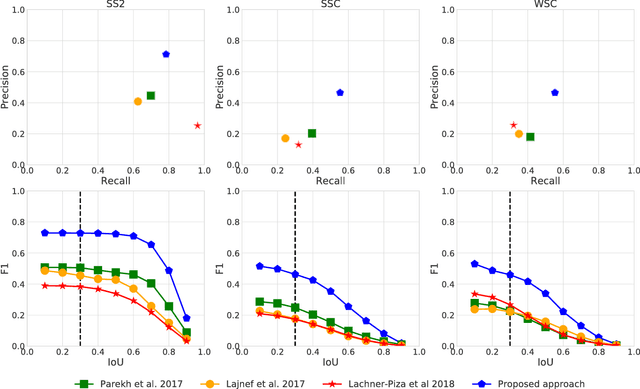
Abstract:Background: Electroencephalography (EEG) monitors brain activity during sleep and is used to identify sleep disorders. In sleep medicine, clinicians interpret raw EEG signals in so-called sleep stages, which are assigned by experts to every 30s window of signal. For diagnosis, they also rely on shorter prototypical micro-architecture events which exhibit variable durations and shapes, such as spindles, K-complexes or arousals. Annotating such events is traditionally performed by a trained sleep expert, making the process time consuming, tedious and subject to inter-scorer variability. To automate this procedure, various methods have been developed, yet these are event-specific and rely on the extraction of hand-crafted features. New method: We propose a novel deep learning architecure called Dreem One Shot Event Detector (DOSED). DOSED jointly predicts locations, durations and types of events in EEG time series. The proposed approach, applied here on sleep related micro-architecture events, is inspired by object detectors developed for computer vision such as YOLO and SSD. It relies on a convolutional neural network that builds a feature representation from raw EEG signals, as well as two modules performing localization and classification respectively. Results and comparison with other methods: The proposed approach is tested on 4 datasets and 3 types of events (spindles, K-complexes, arousals) and compared to the current state-of-the-art detection algorithms. Conclusions: Results demonstrate the versatility of this new approach and improved performance compared to the current state-of-the-art detection methods.
Deep residual networks for automatic sleep stage classification of raw polysomnographic waveforms
Oct 08, 2018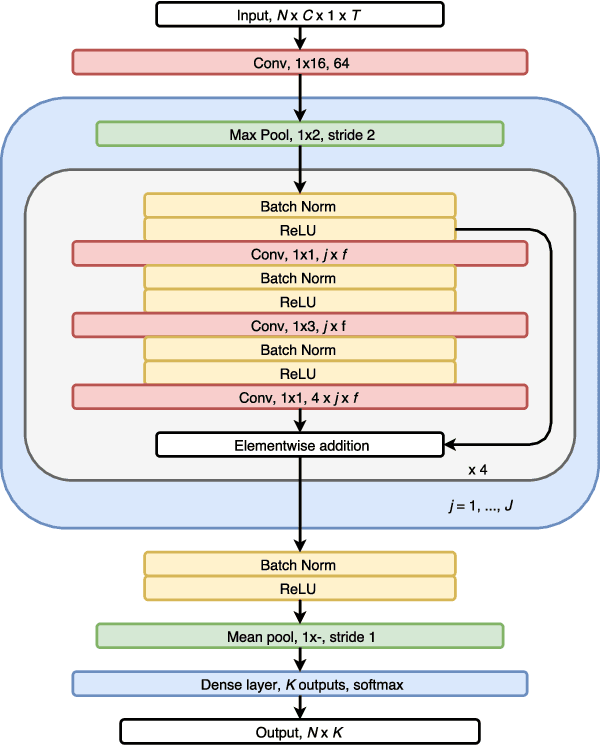
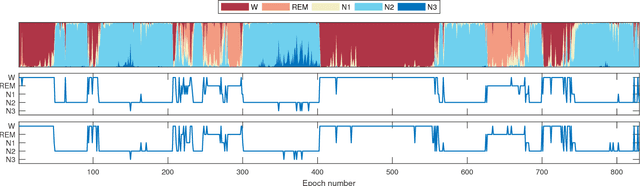
Abstract:We have developed an automatic sleep stage classification algorithm based on deep residual neural networks and raw polysomnogram signals. Briefly, the raw data is passed through 50 convolutional layers before subsequent classification into one of five sleep stages. Three model configurations were trained on 1850 polysomnogram recordings and subsequently tested on 230 independent recordings. Our best performing model yielded an accuracy of 84.1% and a Cohen's kappa of 0.746, improving on previous reported results by other groups also using only raw polysomnogram data. Most errors were made on non-REM stage 1 and 3 decisions, errors likely resulting from the definition of these stages. Further testing on independent cohorts is needed to verify performance for clinical use.
A deep learning architecture to detect events in EEG signals during sleep
Jul 11, 2018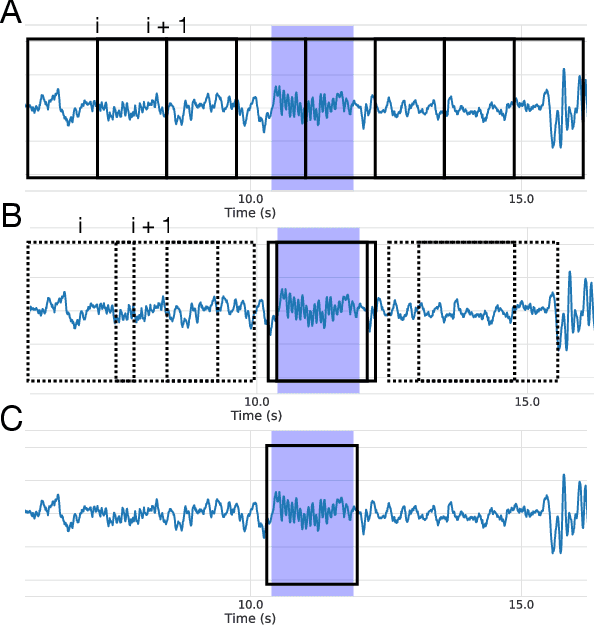
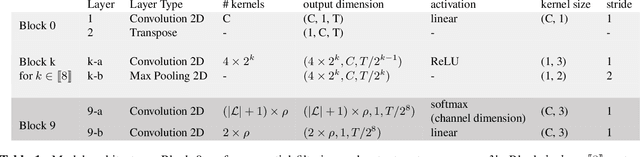
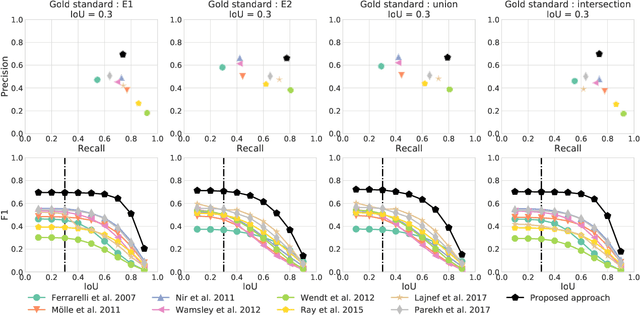
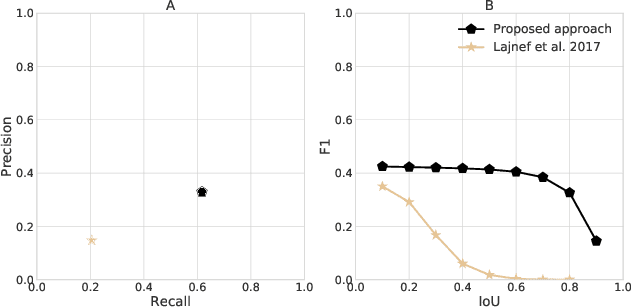
Abstract:Electroencephalography (EEG) during sleep is used by clinicians to evaluate various neurological disorders. In sleep medicine, it is relevant to detect macro-events (> 10s) such as sleep stages, and micro-events (<2s) such as spindles and K-complexes. Annotations of such events require a trained sleep expert, a time consuming and tedious process with a large inter-scorer variability. Automatic algorithms have been developed to detect various types of events but these are event-specific. We propose a deep learning method that jointly predicts locations, durations and types of events in EEG time series. It relies on a convolutional neural network that builds a feature representation from raw EEG signals. Numerical experiments demonstrate efficiency of this new approach on various event detection tasks compared to current state-of-the-art, event specific, algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge