Helge B. D. Sorensen
MSED: a multi-modal sleep event detection model for clinical sleep analysis
Jan 07, 2021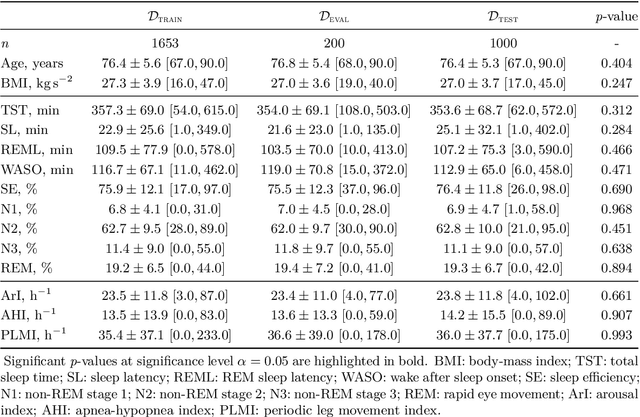
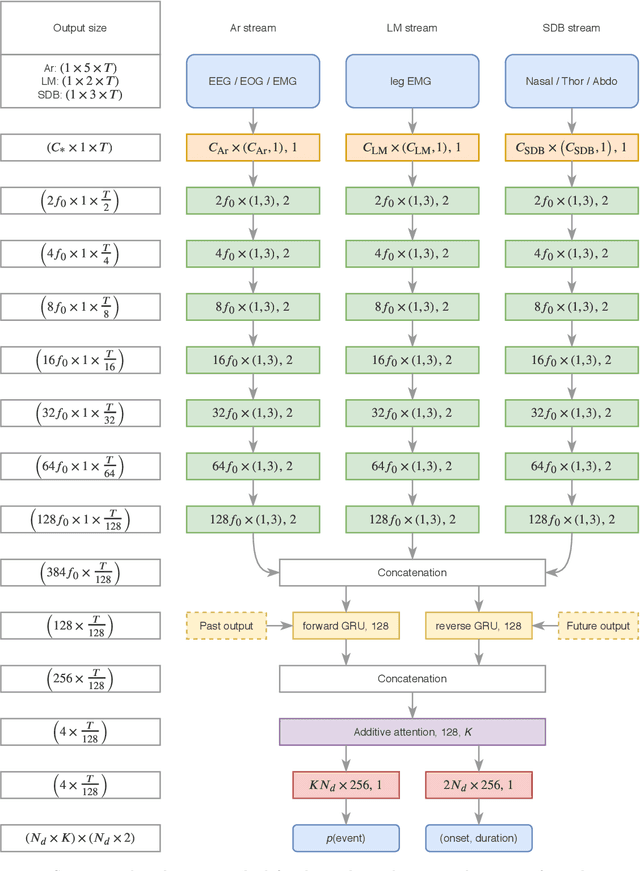
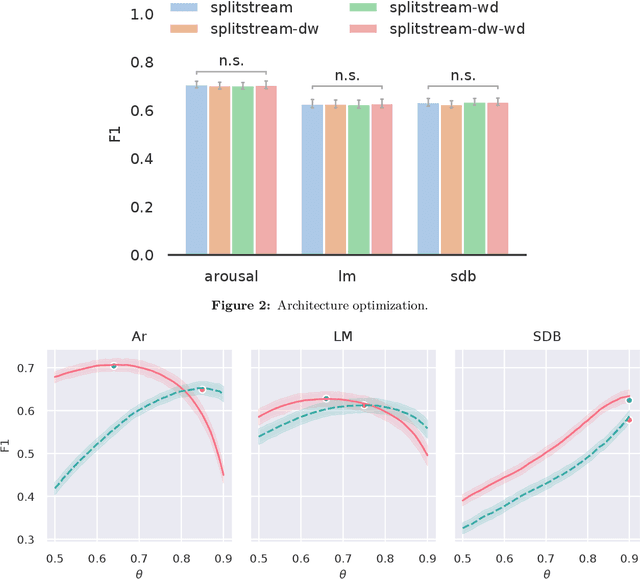
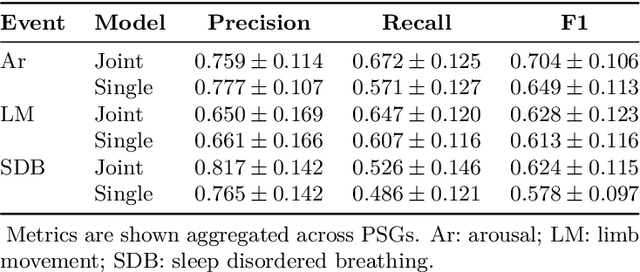
Abstract:Study objective: Clinical sleep analysis require manual analysis of sleep patterns for correct diagnosis of sleep disorders. Several studies show significant variability in scoring discrete sleep events. We wished to investigate, whether an automatic method could be used for detection of arousals (Ar), leg movements (LM) and sleep disordered breathing (SDB) events, and if the joint detection of these events performed better than having three separate models. Methods: We designed a single deep neural network architecture to jointly detect sleep events in a polysomnogram. We trained the model on 1653 recordings of individuals, and tested the optimized model on 1000 separate recordings. The performance of the model was quantified by F1, precision, and recall scores, and by correlating index values to clinical values using Pearson's correlation coefficient. Results: F1 scores for the optimized model was 0.70, 0.63, and 0.62 for Ar, LM, and SDB, respectively. The performance was higher, when detecting events jointly compared to corresponding single-event models. Index values computed from detected events correlated well with manual annotations ($r^2$ = 0.73, $r^2$ = 0.77, $r^2$ = 0.78, respectively). Conclusion: Detecting arousals, leg movements and sleep disordered breathing events jointly is possible, and the computed index values correlates well with human annotations.
Deep transfer learning for improving single-EEG arousal detection
May 07, 2020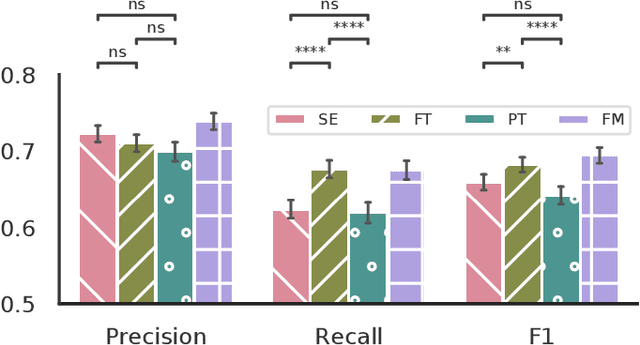
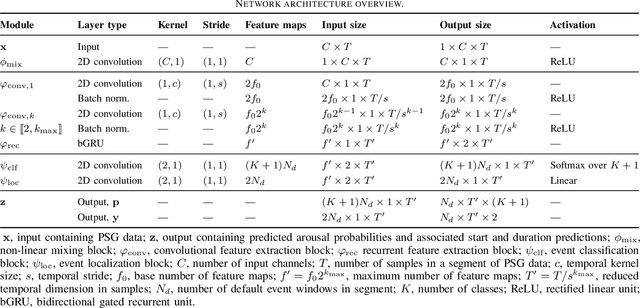
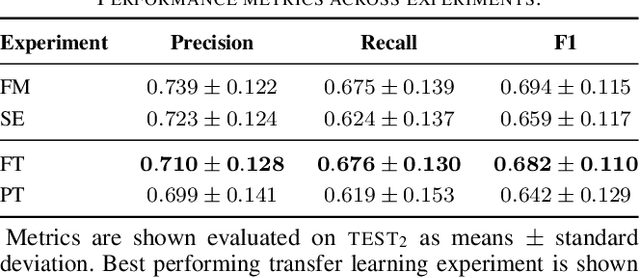
Abstract:Datasets in sleep science present challenges for machine learning algorithms due to differences in recording setups across clinics. We investigate two deep transfer learning strategies for overcoming the channel mismatch problem for cases where two datasets do not contain exactly the same setup leading to degraded performance in single-EEG models. Specifically, we train a baseline model on multivariate polysomnography data and subsequently replace the first two layers to prepare the architecture for single-channel electroencephalography data. Using a fine-tuning strategy, our model yields similar performance to the baseline model (F1=0.682 and F1=0.694, respectively), and was significantly better than a comparable single-channel model. Our results are promising for researchers working with small databases who wish to use deep learning models pre-trained on larger databases.
Towards a Flexible Deep Learning Method for Automatic Detection of Clinically Relevant Multi-Modal Events in the Polysomnogram
May 16, 2019



Abstract:Much attention has been given to automatic sleep staging algorithms in past years, but the detection of discrete events in sleep studies is also crucial for precise characterization of sleep patterns and possible diagnosis of sleep disorders. We propose here a deep learning model for automatic detection and annotation of arousals and leg movements. Both of these are commonly seen during normal sleep, while an excessive amount of either is linked to disrupted sleep patterns, excessive daytime sleepiness impacting quality of life, and various sleep disorders. Our model was trained on 1,485 subjects and tested on 1,000 separate recordings of sleep. We tested two different experimental setups and found optimal arousal detection was attained by including a recurrent neural network module in our default model with a dynamic default event window (F1 = 0.75), while optimal leg movement detection was attained using a static event window (F1 = 0.65). Our work show promise while still allowing for improvements. Specifically, future research will explore the proposed model as a general-purpose sleep analysis model.
The use of neural networks in the analysis of sleep stages and the diagnosis of narcolepsy
Oct 05, 2017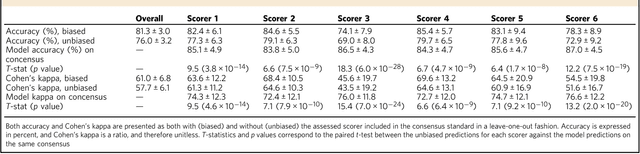
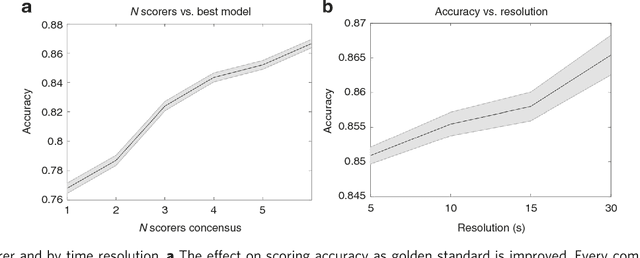

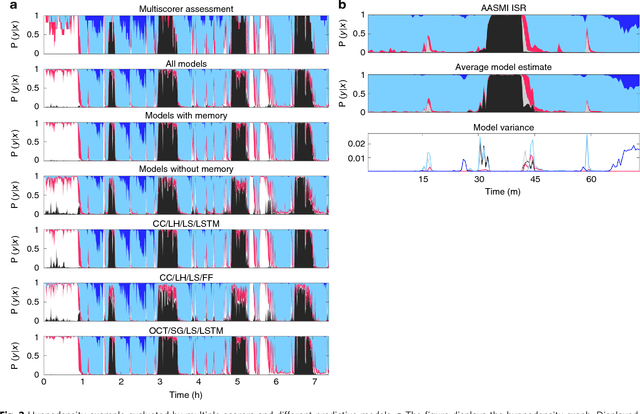
Abstract:We used neural networks in ~3,000 sleep recordings from over 10 locations to automate sleep stage scoring, producing a probability distribution called an hypnodensity graph. Accuracy was validated in 70 subjects scored by six technicians (gold standard). Our best model performed better than any individual scorer, reaching an accuracy of 0.87 (and 0.95 when predictions are weighed by scorer agreement). It also scores sleep stages down to 5-second instead of the conventional 30-second scoring-epochs. Accuracy did not vary by sleep disorder except for narcolepsy, suggesting scoring difficulties by machine and/or humans. A narcolepsy biomarker was extracted and validated in 105 type-1 narcoleptics versus 331 controls producing a specificity of 0.96 and a sensitivity of 0.91. Similar performances were obtained against a high pretest probability sample of type-2 narcolepsy and idiopathic hypersomnia patients. Addition of HLA-DQB1*06:02 increased specificity to 0.99. Our method streamlines scoring and diagnoses narcolepsy accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge