Stanislas Chambon
Towards a Flexible Deep Learning Method for Automatic Detection of Clinically Relevant Multi-Modal Events in the Polysomnogram
May 16, 2019



Abstract:Much attention has been given to automatic sleep staging algorithms in past years, but the detection of discrete events in sleep studies is also crucial for precise characterization of sleep patterns and possible diagnosis of sleep disorders. We propose here a deep learning model for automatic detection and annotation of arousals and leg movements. Both of these are commonly seen during normal sleep, while an excessive amount of either is linked to disrupted sleep patterns, excessive daytime sleepiness impacting quality of life, and various sleep disorders. Our model was trained on 1,485 subjects and tested on 1,000 separate recordings of sleep. We tested two different experimental setups and found optimal arousal detection was attained by including a recurrent neural network module in our default model with a dynamic default event window (F1 = 0.75), while optimal leg movement detection was attained using a static event window (F1 = 0.65). Our work show promise while still allowing for improvements. Specifically, future research will explore the proposed model as a general-purpose sleep analysis model.
DOSED: a deep learning approach to detect multiple sleep micro-events in EEG signal
Dec 07, 2018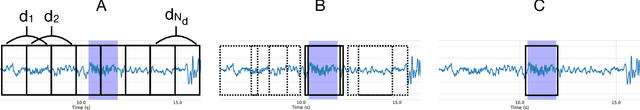
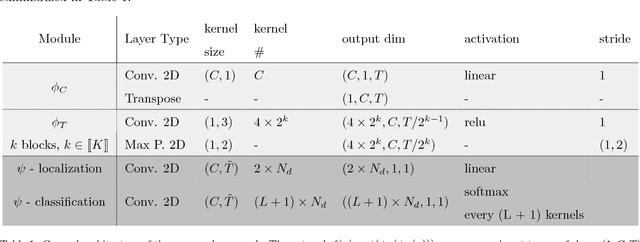
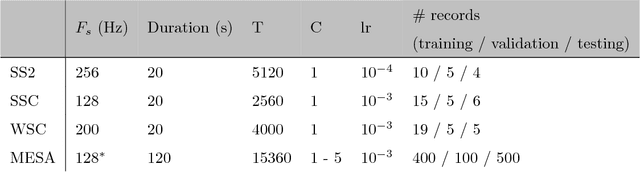
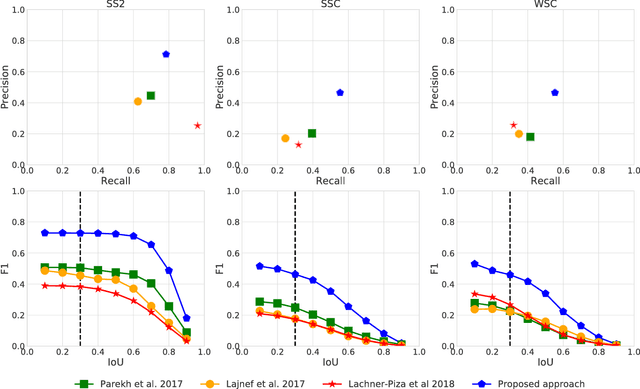
Abstract:Background: Electroencephalography (EEG) monitors brain activity during sleep and is used to identify sleep disorders. In sleep medicine, clinicians interpret raw EEG signals in so-called sleep stages, which are assigned by experts to every 30s window of signal. For diagnosis, they also rely on shorter prototypical micro-architecture events which exhibit variable durations and shapes, such as spindles, K-complexes or arousals. Annotating such events is traditionally performed by a trained sleep expert, making the process time consuming, tedious and subject to inter-scorer variability. To automate this procedure, various methods have been developed, yet these are event-specific and rely on the extraction of hand-crafted features. New method: We propose a novel deep learning architecure called Dreem One Shot Event Detector (DOSED). DOSED jointly predicts locations, durations and types of events in EEG time series. The proposed approach, applied here on sleep related micro-architecture events, is inspired by object detectors developed for computer vision such as YOLO and SSD. It relies on a convolutional neural network that builds a feature representation from raw EEG signals, as well as two modules performing localization and classification respectively. Results and comparison with other methods: The proposed approach is tested on 4 datasets and 3 types of events (spindles, K-complexes, arousals) and compared to the current state-of-the-art detection algorithms. Conclusions: Results demonstrate the versatility of this new approach and improved performance compared to the current state-of-the-art detection methods.
A deep learning architecture to detect events in EEG signals during sleep
Jul 11, 2018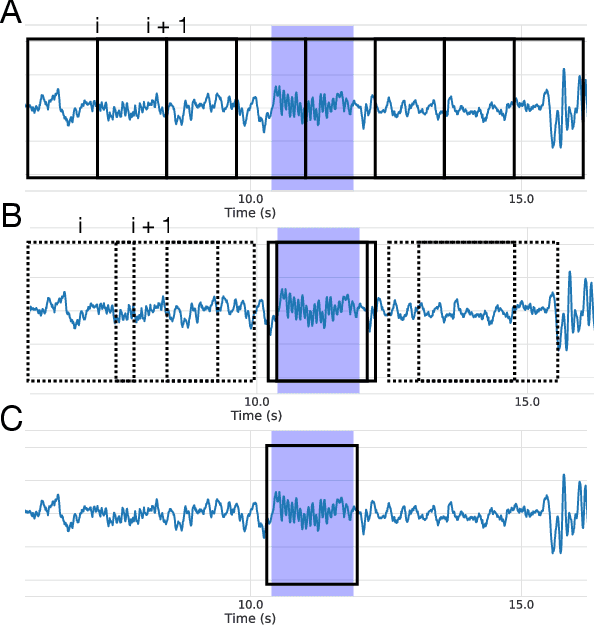
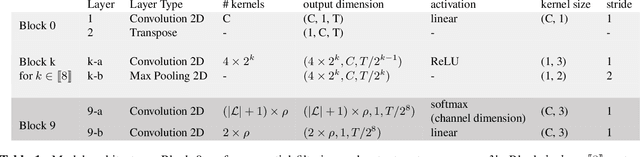
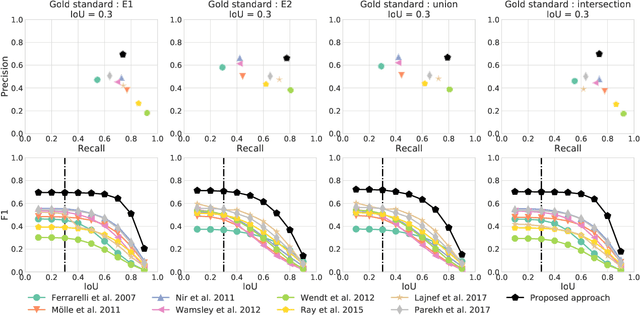
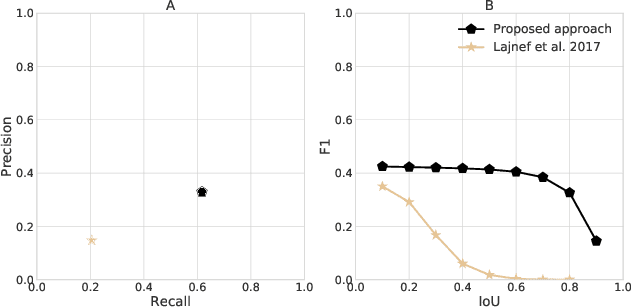
Abstract:Electroencephalography (EEG) during sleep is used by clinicians to evaluate various neurological disorders. In sleep medicine, it is relevant to detect macro-events (> 10s) such as sleep stages, and micro-events (<2s) such as spindles and K-complexes. Annotations of such events require a trained sleep expert, a time consuming and tedious process with a large inter-scorer variability. Automatic algorithms have been developed to detect various types of events but these are event-specific. We propose a deep learning method that jointly predicts locations, durations and types of events in EEG time series. It relies on a convolutional neural network that builds a feature representation from raw EEG signals. Numerical experiments demonstrate efficiency of this new approach on various event detection tasks compared to current state-of-the-art, event specific, algorithms.
A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series
Nov 27, 2017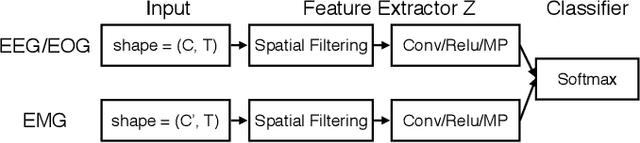
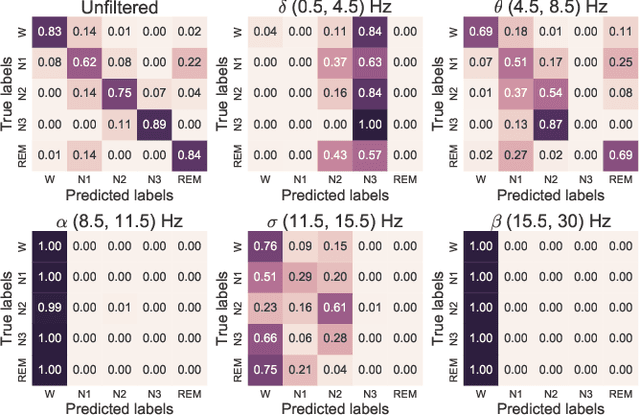
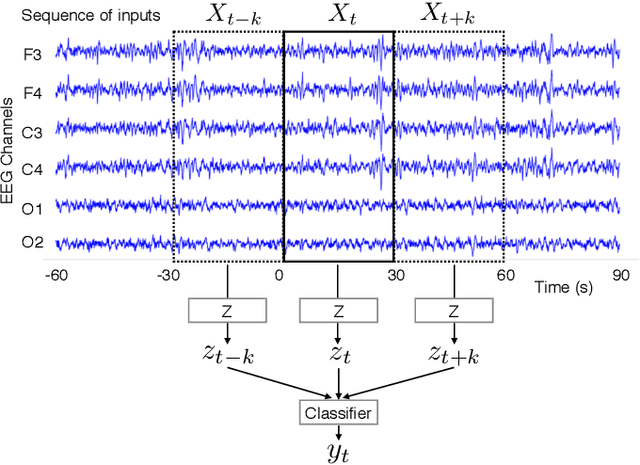
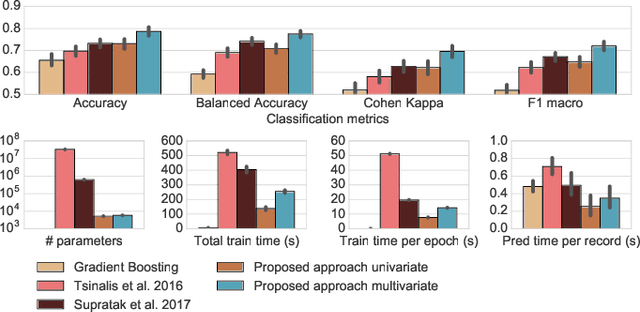
Abstract:Sleep stage classification constitutes an important preliminary exam in the diagnosis of sleep disorders. It is traditionally performed by a sleep expert who assigns to each 30s of signal a sleep stage, based on the visual inspection of signals such as electroencephalograms (EEG), electrooculograms (EOG), electrocardiograms (ECG) and electromyograms (EMG). We introduce here the first deep learning approach for sleep stage classification that learns end-to-end without computing spectrograms or extracting hand-crafted features, that exploits all multivariate and multimodal Polysomnography (PSG) signals (EEG, EMG and EOG), and that can exploit the temporal context of each 30s window of data. For each modality the first layer learns linear spatial filters that exploit the array of sensors to increase the signal-to-noise ratio, and the last layer feeds the learnt representation to a softmax classifier. Our model is compared to alternative automatic approaches based on convolutional networks or decisions trees. Results obtained on 61 publicly available PSG records with up to 20 EEG channels demonstrate that our network architecture yields state-of-the-art performance. Our study reveals a number of insights on the spatio-temporal distribution of the signal of interest: a good trade-off for optimal classification performance measured with balanced accuracy is to use 6 EEG with 2 EOG (left and right) and 3 EMG chin channels. Also exploiting one minute of data before and after each data segment offers the strongest improvement when a limited number of channels is available. As sleep experts, our system exploits the multivariate and multimodal nature of PSG signals in order to deliver state-of-the-art classification performance with a small computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge